5CVC
 
 | | Structure of maize serine racemase | | Descriptor: | MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, Serine racemase | | Authors: | Song, Y, Zou, L, Fan, J. | | Deposit date: | 2015-07-26 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of maize serine racemase with pyridoxal 5'-phosphate.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
8QBG
 
 | |
8QBF
 
 | | Compact state - Pil1 dimer with lipid headgroups fitted in native eisosome lattice bound to plasma membrane microdomain | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PHOSPHOSERINE, Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | CryoEM architecture of a native stretch-sensitive membrane microdomain
To Be Published
|
|
8QB9
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture -PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol 30:20:20:30) | | Descriptor: | Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | CryoEM architecture of a native stretch-sensitive membrane microdomain
To Be Published
|
|
8QBD
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol, PI(4,5)P2 35:20:20:15:10) | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | CryoEM architecture of a native stretch-sensitive membrane microdomain
To Be Published
|
|
8QB8
 
 | |
8QB7
 
 | |
8QBE
 
 | |
8QBB
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/-sterol (DOPC, DOPE, DOPS, PI(4,5)P2 50:20:20:10) | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | CryoEM architecture of a native stretch-sensitive membrane microdomain
To Be Published
|
|
3N3Z
 
 | | Crystal structure of PDE9A (E406A) mutant in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, CHLORIDE ION, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, ... | | Authors: | Hou, J, Luo, H.-B, Chen, Y, Xu, J, Zhao, R, Zou, L. | | Deposit date: | 2010-05-21 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of PDE9A (E406A) mutation in complex with IBMX
To be Published
|
|
7XNE
 
 | | Crystal structure of CBP bromodomain liganded with Y08284 | | Descriptor: | CREB-binding protein, GLYCEROL, N-[3-(1-cyclopropylpyrazol-4-yl)-2-fluoranyl-5-[(1S)-1-oxidanylethyl]phenyl]-3-ethanoyl-7-methoxy-indolizine-1-carboxamide | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, C, Hu, Q, Luo, G, Hu, J, Zhuang, X, Zou, L, Shen, H, Wu, X, Zhang, Y, Kong, X, Xu, Y. | | Deposit date: | 2022-04-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
1Z1D
 
 | | Structural Model for the interaction between RPA32 C-terminal domain and SV40 T antigen origin binding domain. | | Descriptor: | Large T antigen, Replication protein A 32 kDa subunit | | Authors: | Arunkumar, A.I, Klimovich, V, Jiang, X, Ott, R.D, Mizoue, L, Fanning, E, Chazin, W.J. | | Deposit date: | 2005-03-03 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Insights into hRPA32 C-terminal domain--mediated assembly of the simian virus 40 replisome.
Nat.Struct.Mol.Biol., 12, 2005
|
|
3TDM
 
 | | Computationally designed TIM-barrel protein, HalfFLR | | Descriptor: | Computationally designed two-fold symmetric TIM-barrel protein, FLR (half molecule), PHOSPHATE ION | | Authors: | Harp, J.M, Fortenberry, C, Bowman, E, Profitt, W, Dorr, B, Mizoue, L. | | Deposit date: | 2011-08-11 | | Release date: | 2011-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploring symmetry as an avenue to the computational design of large protein domains.
J.Am.Chem.Soc., 133, 2011
|
|
3TDN
 
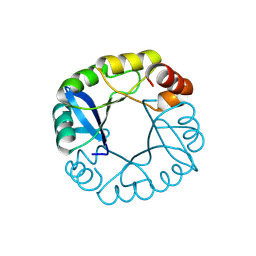 | | Computationally designed two-fold symmetric Tim-barrel protein, FLR | | Descriptor: | FLR SYMMETRIC ALPHA-BETA TIM BARREL | | Authors: | Harp, J.M, Fortenberry, C, Bowman, E, Profitt, W, Dorr, B, Mizoue, L, Meiler, J. | | Deposit date: | 2011-08-11 | | Release date: | 2011-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Exploring symmetry as an avenue to the computational design of large protein domains.
J.Am.Chem.Soc., 133, 2011
|
|
6J1L
 
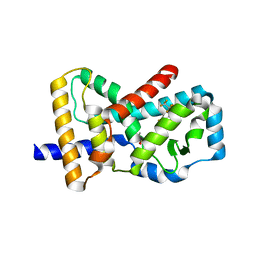 | | Crystal Structure Analysis of the ROR gamma(C455E) | | Descriptor: | 2-[4-(ethylsulfonyl)phenyl]-N-[2'-fluoro-4'-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)[1,1'-biphenyl]-4-yl]acetamide, Nuclear receptor ROR-gamma | | Authors: | zhang, Y, Li, C.C, wu, X.S. | | Deposit date: | 2018-12-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Characterization of XY101, a Potent, Selective, and Orally Bioavailable ROR gamma Inverse Agonist for Treatment of Castration-Resistant Prostate Cancer.
J.Med.Chem., 62, 2019
|
|
7VZF
 
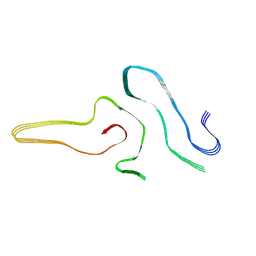 | | Cryo-EM structure of amyloid fibril formed by full-length human SOD1 | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Wang, L.Q, Ma, Y.Y, Yuan, H.Y, Zhao, K, Zhang, M.Y, Wang, Q, Huang, X, Xu, W.C, Chen, J, Li, D, Zhang, D.L, Zou, L.Y, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structure of an amyloid fibril formed by full-length human SOD1 reveals its conformational conversion.
Nat Commun, 13, 2022
|
|
5Y9Q
 
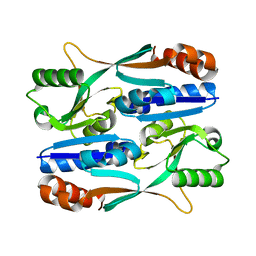 | |
6A8L
 
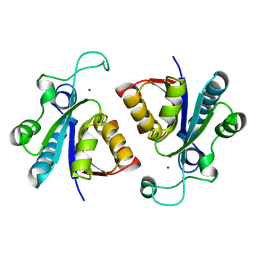 | | Crystal structure of nicotinamidase/ pyrazinamidase PncA from Bacillus subtilis | | Descriptor: | Isochorismatase, ZINC ION | | Authors: | Shang, F, Chen, J, Wang, L, Xu, Y. | | Deposit date: | 2018-07-09 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the nicotinamidase/pyrazinamidase PncA from Bacillus subtilis.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
5ZZO
 
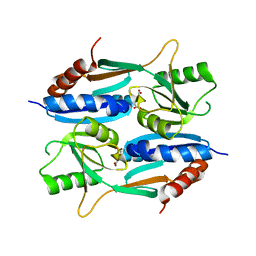 | | Crystal structure of CcpE regulatory domain in complex with citrate from Staphyloccocus aureus | | Descriptor: | CITRATE ANION, LysR family transcriptional regulator | | Authors: | Chen, J, Wang, L, Shang, F, Xu, Y. | | Deposit date: | 2018-06-04 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Analysis of the Citrate-Responsive Mechanism of the Regulatory Domain of Catabolite Control Protein E from Staphylococcus aureus
Biochemistry, 57, 2018
|
|
5ZN8
 
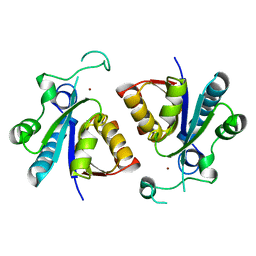 | | Crystal structure of nicotinamidase PncA from Bacillus subtilis | | Descriptor: | Isochorismatase, ZINC ION | | Authors: | Shang, F, Chen, J, Wang, L, Xu, Y. | | Deposit date: | 2018-04-08 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the nicotinamidase/pyrazinamidase PncA from Bacillus subtilis.
Biochem.Biophys.Res.Commun., 503, 2018
|
|
8R1L
 
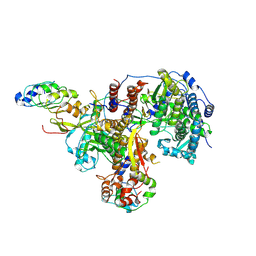 | | Structure of avian H5N1 influenza A polymerase in complex with human ANP32B. | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Staller, E, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2023-11-02 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | CryoEM architecture of a native stretch-sensitive membrane microdomain
To Be Published
|
|
5Y8C
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloranyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y93
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[5-[(5-bromanyl-2-methoxy-phenyl)sulfonylamino]-3-methyl-1,2-benzoxazol-6-yl]oxy]-N-(2-morpholin-4-ylethyl)ethanamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y8W
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-N-(3-methyl-6-oxidanyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y94
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 5-bromanyl-2-methoxy-N-[3-methyl-6-(methylamino)-1,2-benzoxazol-5-yl]benzenesulfonamide, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
