4I4Z
 
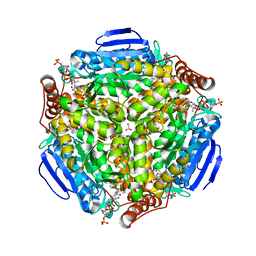 | | Synechocystis sp. PCC 6803 1,4-dihydroxy-2-naphthoyl-coenzyme A synthase (MenB) in complex with salicylyl-CoA | | Descriptor: | BICARBONATE ION, MALONATE ION, Naphthoate synthase, ... | | Authors: | Song, H.G, Sun, Y.R, Li, J, Li, Y, Jiang, M, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme a synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
4I52
 
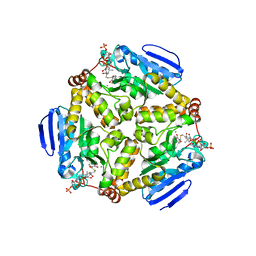 | | scMenB im complex with 1-hydroxy-2-naphthoyl-CoA | | Descriptor: | 1-hydroxy-2-naphthoyl-CoA, CHLORIDE ION, Naphthoate synthase | | Authors: | Song, H.G, Sun, Y.R, Li, J, Li, Y, Jiang, M, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme a synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
2QP9
 
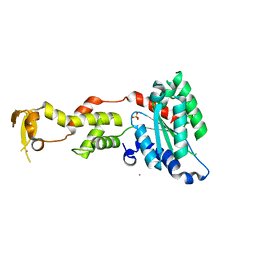 | | Crystal Structure of S.cerevisiae Vps4 | | Descriptor: | CADMIUM ION, SULFATE ION, Vacuolar protein sorting-associated protein 4 | | Authors: | Xiao, J, Xu, Z. | | Deposit date: | 2007-07-23 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural characterization of the ATPase reaction cycle of endosomal AAA protein Vps4.
J.Mol.Biol., 374, 2007
|
|
8DB4
 
 | |
4O98
 
 | | Crystal structure of Pseudomonas oleovorans PoOPH mutant H250I/I263W | | Descriptor: | ZINC ION, organophosphorus hydrolase | | Authors: | Luo, X.J, Kong, X.D, Zhao, J, Chen, Q, Zhou, J.H, Xu, J.H. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-03 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Switching a newly discovered lactonase into an efficient and thermostable phosphotriesterase by simple double mutations His250Ile/Ile263Trp
Biotechnol.Bioeng., 111, 2014
|
|
8GT6
 
 | | human STING With agonist HB3089 | | Descriptor: | 1-[(2E)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methyl-1H-furo[3,2-e]benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
8GSZ
 
 | | Structure of STING SAVI-related mutant V147L | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
7C53
 
 | |
7ASL
 
 | |
7ASH
 
 | |
4ERF
 
 | | crystal structure of MDM2 (17-111) in complex with compound 29 (AM-8553) | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-[(2S,3S)-2-hydroxypentan-3-yl]-3-methyl-2-oxopiperidin-3-yl}acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
4ERE
 
 | | crystal structure of MDM2 (17-111) in complex with compound 23 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(3R,5R,6S)-1-[(2S)-1-tert-butoxy-1-oxobutan-2-yl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-2-oxopiperidin-3-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
8G05
 
 | | Cryo-EM structure of an orphan GPCR-Gi protein signaling complex | | Descriptor: | 6-(octylamino)pyrimidine-2,4(3H,5H)-dione, CHOLESTEROL, G-protein coupled receptor 84, ... | | Authors: | Zhang, X, Wang, Y.J, Li, X, Liu, G.B, Gong, W.M, Zhang, C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Pro-phagocytic function and structural basis of GPR84 signaling.
Nat Commun, 14, 2023
|
|
7UCC
 
 | | Transcription factor FosB/JunD bZIP domain in the reduced form | | Descriptor: | CHLORIDE ION, ETHANOL, Protein fosB, ... | | Authors: | Kumar, A, Machius, M.C, Rudenko, G. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Chemically targeting the redox switch in AP1 transcription factor Delta FOSB.
Nucleic Acids Res., 50, 2022
|
|
7UCD
 
 | | Transcription factor FosB/JunD bZIP domain covalently modified with the cysteine-targeting alpha-haloketone compound Z2159931480 | | Descriptor: | 7-acetyl-4-methoxy-1-benzofuran-3(2H)-one, CHLORIDE ION, Protein fosB, ... | | Authors: | Kumar, A, Machius, M.C, Rudenko, G. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Chemically targeting the redox switch in AP1 transcription factor Delta FOSB.
Nucleic Acids Res., 50, 2022
|
|
7CSD
 
 | | AtPrR1 with NADP+ and (+)lariciresinol | | Descriptor: | 4-[[(3R,4R,5S)-4-(hydroxymethyl)-5-(3-methoxy-4-oxidanyl-phenyl)oxolan-3-yl]methyl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79709971 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS2
 
 | | Apo structure of dimeric IiPLR1 | | Descriptor: | Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68800473 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS6
 
 | | IiPLR1 with NADP+ and (-)lariciresinol | | Descriptor: | 4-[[(3S,4S,5R)-4-(hydroxymethyl)-5-(3-methoxy-4-oxidanyl-phenyl)oxolan-3-yl]methyl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20139647 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS8
 
 | | IiPLR1 with NADP+ and (-)secoisolariciresinol | | Descriptor: | (2R,3R)-2,3-bis[(3-methoxy-4-oxidanyl-phenyl)methyl]butane-1,4-diol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.60003257 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSF
 
 | | AtPrR1 with NADP+ and (-)secoisolariciresinol | | Descriptor: | (2R,3R)-2,3-bis[(3-methoxy-4-oxidanyl-phenyl)methyl]butane-1,4-diol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98241806 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS3
 
 | | IiPLR1 with NADP+ | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.40021849 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSE
 
 | | AtPrR1 with NADP+ and (-)lariciresinol | | Descriptor: | 4-[[(3S,4S,5R)-4-(hydroxymethyl)-5-(3-methoxy-4-oxidanyl-phenyl)oxolan-3-yl]methyl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44019055 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSC
 
 | | AtPrR1 with NADP+ and (-)pinoresinol | | Descriptor: | 4-[(3R,3aS,6R,6aS)-6-(3-methoxy-4-oxidanyl-phenyl)-1,3,3a,4,6,6a-hexahydrofuro[3,4-c]furan-3-yl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5151186 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS4
 
 | | IiPLR1 with NADP+ and (+)pinoresinol | | Descriptor: | 4-[(3S,3aR,6S,6aR)-6-(3-methoxy-4-oxidanyl-phenyl)-1,3,3a,4,6,6a-hexahydrofuro[3,4-c]furan-3-yl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.30509257 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS5
 
 | | IiPLR1 with NADP+ and (-)pinoresinol | | Descriptor: | 4-[(3R,3aS,6R,6aS)-6-(3-methoxy-4-oxidanyl-phenyl)-1,3,3a,4,6,6a-hexahydrofuro[3,4-c]furan-3-yl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18993235 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
