7DJY
 
 | | Clec4f Nanobody 322 | | Descriptor: | Clec4f Nanobody 322 | | Authors: | Wen, Y, Zheng, F. | | Deposit date: | 2020-11-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Clec4f Nanobody 322
To Be Published
|
|
6YK0
 
 | |
6YJZ
 
 | |
6YK1
 
 | |
3ICH
 
 | |
3ICI
 
 | |
6W07
 
 | | Bruton's tyrosine kinase in complex with compound 1 | | Descriptor: | DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK, ~{N}-[[2-methyl-4-[2-[(1-methylpyrazol-4-yl)amino]pyrimidin-4-yl]phenyl]methyl]-3-propan-2-yloxy-azetidine-1-carboxamide | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Discovery of BIIB068: A Selective, Potent, Reversible Bruton's Tyrosine Kinase Inhibitor as an Orally Efficacious Agent for Autoimmune Diseases.
J.Med.Chem., 63, 2020
|
|
6VXQ
 
 | | Bruton's tyrosine kinase in complex with compound 5 | | Descriptor: | DIMETHYL SULFOXIDE, N-{[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)phenyl]methyl}benzamide, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2020-02-23 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of BIIB068: A Selective, Potent, Reversible Bruton's Tyrosine Kinase Inhibitor as an Orally Efficacious Agent for Autoimmune Diseases.
J.Med.Chem., 63, 2020
|
|
6W06
 
 | | Bruton's tyrosine kinase in complex with compound 6 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-~{tert}-butyl-~{N}-[[4-(7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)phenyl]methyl]benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of BIIB068: A Selective, Potent, Reversible Bruton's Tyrosine Kinase Inhibitor as an Orally Efficacious Agent for Autoimmune Diseases.
J.Med.Chem., 63, 2020
|
|
5IML
 
 | | Nanobody targeting human Vsig4 in Spacegroup P212121 | | Descriptor: | Nanobody, V-set and immunoglobulin domain-containing protein 4 | | Authors: | Wen, Y. | | Deposit date: | 2016-03-06 | | Release date: | 2017-01-11 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity
Immunobiology, 222, 2017
|
|
5IMK
 
 | | Nanobody targeting human Vsig4 in Spacegroup C2 | | Descriptor: | Nanobody, V-set and immunoglobulin domain-containing protein 4 | | Authors: | Wen, Y. | | Deposit date: | 2016-03-06 | | Release date: | 2017-01-11 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.227 Å) | | Cite: | Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity
Immunobiology, 222, 2017
|
|
8H3Y
 
 | | Bacteroide Fragilis Toxin in complex with nanobody 327 | | Descriptor: | Fragilysin, Nanobody 327, ZINC ION | | Authors: | Wen, Y, Guo, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Screening and epitope characterization of diagnostic nanobody against total and activated Bacteroides fragilis toxin.
Front Immunol, 14, 2023
|
|
8H3X
 
 | | Bacteroide Fragilis Toxin in complex with nanobody 282 | | Descriptor: | Fragilysin, ZINC ION, nanobody 282 | | Authors: | Wen, Y, Guo, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Screening and epitope characterization of diagnostic nanobody against total and activated Bacteroides fragilis toxin.
Front Immunol, 14, 2023
|
|
8GRA
 
 | |
5WWL
 
 | | Crystal structure of the Schizogenesis pombe kinetochore Mis12C subcomplex | | Descriptor: | Centromere protein mis12, Kinetochore protein nnf1 | | Authors: | Wang, C, Zhou, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2017-01-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphorylation of CENP-C by Aurora B facilitates kinetochore attachment error correction in mitosis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4P08
 
 | | Engineered thermostable dimeric cocaine esterase | | Descriptor: | Cocaine esterase | | Authors: | Rodgers, D.W, Chow, K.-M, Fang, L, Zhan, C.-G. | | Deposit date: | 2014-02-20 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Rational design, preparation, and characterization of a therapeutic enzyme mutant with improved stability and function for cocaine detoxification.
Acs Chem.Biol., 9, 2014
|
|
5X5L
 
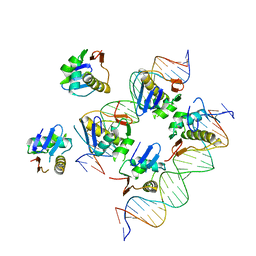 | | Crystal structure of response regulator AdeR DNA binding domain in complex with an intercistronic region | | Descriptor: | AdeR, DNA (5'-D(P*TP*AP*AP*AP*GP*TP*GP*TP*GP*GP*AP*GP*TP*AP*AP*GP*TP*GP*TP*GP*GP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*CP*CP*AP*CP*AP*CP*TP*TP*AP*CP*TP*CP*CP*AP*CP*AP*CP*TP*TP*TP*A)-3') | | Authors: | Wen, Y. | | Deposit date: | 2017-02-16 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region.
Nucleic Acids Res., 45, 2017
|
|
5X5J
 
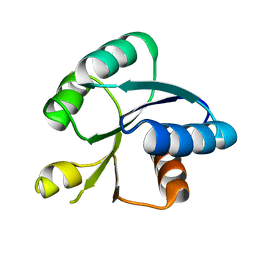 | | Crystal structure of response regulator AdeR receiver domain | | Descriptor: | AdeR | | Authors: | Wen, Y. | | Deposit date: | 2017-02-16 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region.
Nucleic Acids Res., 45, 2017
|
|
5XJP
 
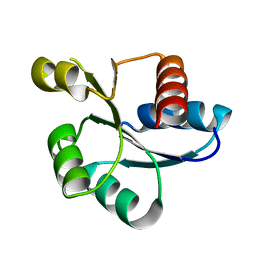 | | Crystal structure of response regulator AdeR receiver domain with Mg | | Descriptor: | AdeR, MAGNESIUM ION | | Authors: | Wen, Y. | | Deposit date: | 2017-05-03 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region.
Nucleic Acids Res., 45, 2017
|
|
5XP0
 
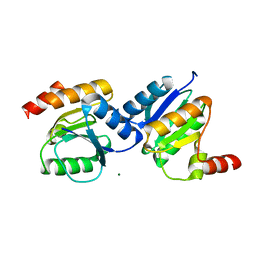 | |
6IS2
 
 | |
6IS3
 
 | |
6IS1
 
 | |
6IS4
 
 | |
6JJJ
 
 | |
