6DUV
 
 | | Crystal structure of the second bromodomain of human BRD4 in complex with MS417 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, methyl [(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetate | | Authors: | Babault, N, Zhou, M.M. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of Bound Water Molecules in Differential Recognition of Di-Acetylated Histone Peptides by the BRD4 Bromodomains
To be published
|
|
1M3G
 
 | |
5L01
 
 | | Tryptophan 5-hydroxylase in complex with inhibitor (3~{S})-8-[2-azanyl-6-[(1~{R})-1-(4-chloranyl-2-phenyl-phenyl)-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]-2,8-diazaspiro[4.5]decane-3-carboxylic acid | | Descriptor: | (3~{S})-8-[2-azanyl-6-[(1~{R})-1-(4-chloranyl-2-phenyl-phenyl)-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]-2,8-diazaspiro[4.5]decane-3-carboxylic acid, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Stein, A.J, Goldberg, D.R, De Lombaert, S, Holt, M.C. | | Deposit date: | 2016-07-26 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of spirocyclic proline tryptophan hydroxylase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5L1I
 
 | |
5L1L
 
 | | PostInsertion complex of Human DNA Polymerase Eta bypassing an O6-Methyl-2'-deoxyguanosine : dT site | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*GP*(6OG)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mechanisms of Insertion of dCTP and dTTP Opposite the DNA Lesion O6-Methyl-2'-deoxyguanosine by Human DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
5L1K
 
 | | PostInsertion complex of Human DNA Polymerase Eta bypassing an O6-Methyl-2'-deoxyguanosine : dC site | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*C)-3'), DNA (5'-D(*CP*AP*TP*GP*(6OG)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Mechanisms of Insertion of dCTP and dTTP Opposite the DNA Lesion O6-Methyl-2'-deoxyguanosine by Human DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
5L1J
 
 | |
5KQE
 
 | |
5LQ3
 
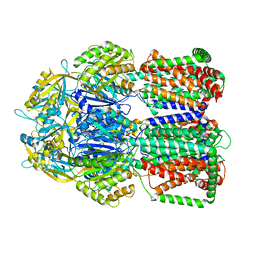 | |
3CIB
 
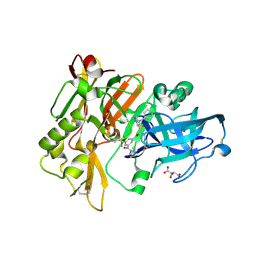 | | Structure of BACE Bound to SCH727596 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2R)-2-[(2R,4S)-4-benzylpiperidin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CID
 
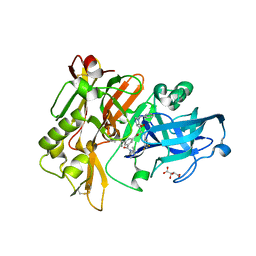 | | Structure of BACE Bound to SCH726222 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(4S)-1-benzyl-5-oxoimidazolidin-4-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5MZ3
 
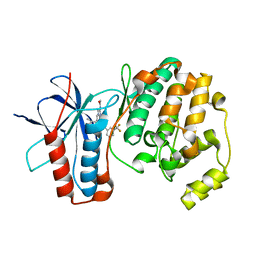 | | P38 ALPHA MUTANT C162S IN COMPLEX WITH CMPD2 [N-(4-Methyl-3-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)-3-(trifluoromethyl)benzamide] | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[3-(2-acetamidoimidazo[1,2-a]pyridin-6-yl)-4-methyl-phenyl]-3-(trifluoromethyl)benzamide | | Authors: | Cowan-Jacob, S.W, Scheufler, C. | | Deposit date: | 2017-01-30 | | Release date: | 2017-11-15 | | Last modified: | 2018-11-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Imidazo[1,2-a]pyridin-6-yl-benzamide analogs as potent RAF inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3CIC
 
 | | Structure of BACE Bound to SCH709583 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(2S)-4-benzyl-3-oxopiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7FJO
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with three T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7FJN
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with two T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7FJS
 
 | | Crystal structure of T6 Fab bound to theSARS-CoV-2 RBD of B.1.351 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
1YW8
 
 | | h-MetAP2 complexed with A751277 | | Descriptor: | 2-[(PHENYLSULFONYL)AMINO]-5,6,7,8-TETRAHYDRONAPHTHALENE-1-CARBOXYLIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2005-02-17 | | Release date: | 2006-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and optimization of anthranilic acid sulfonamides as inhibitors of methionine aminopeptidase-2: a structural basis for the reduction of albumin binding.
J.Med.Chem., 49, 2006
|
|
1YW7
 
 | | h-MetAP2 complexed with A444148 | | Descriptor: | 5-METHYL-2-[(PHENYLSULFONYL)AMINO]BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2005-02-17 | | Release date: | 2006-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and optimization of anthranilic acid sulfonamides as inhibitors of methionine aminopeptidase-2: a structural basis for the reduction of albumin binding.
J.Med.Chem., 49, 2006
|
|
8IQ1
 
 | | Crystal structure of hydrogen sulfide-bound superoxide dismutase in reduced state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Zhou, J.H, Huang, W.X, Cheng, R.X, Zhang, P.J, Zhu, Y.C. | | Deposit date: | 2023-03-15 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydrogen sulfide functions as a micro-modulator bound at the copper active site of Cu/Zn-SOD to regulate the catalytic activity of the enzyme.
Cell Rep, 42, 2023
|
|
8IQ0
 
 | | Crystal structure of hydrogen sulfide-bound superoxide dismutase in oxidized state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Zhou, J.H, Huang, W.X, Cheng, R.X, Zhang, P.J, Zhu, Y.C. | | Deposit date: | 2023-03-15 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Hydrogen sulfide functions as a micro-modulator bound at the copper active site of Cu/Zn-SOD to regulate the catalytic activity of the enzyme.
Cell Rep, 42, 2023
|
|
1YW9
 
 | | h-MetAP2 complexed with A849519 | | Descriptor: | 2-[({2-[(1Z)-3-(DIMETHYLAMINO)PROP-1-ENYL]-4-FLUOROPHENYL}SULFONYL)AMINO]-5,6,7,8-TETRAHYDRONAPHTHALENE-1-CARBOXYLIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2005-02-17 | | Release date: | 2006-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery and optimization of anthranilic acid sulfonamides as inhibitors of methionine aminopeptidase-2: a structural basis for the reduction of albumin binding.
J.Med.Chem., 49, 2006
|
|
1WWN
 
 | | NMR Solution Structure of BmK-betaIT, an Excitatory Scorpion Toxin from Buthus martensi Karsch | | Descriptor: | Excitatory insect selective toxin 1 | | Authors: | Wu, H, Tong, X, Chen, X, Zhang, Q, Zheng, X, Zhang, N, Wu, G. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-17 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of BmK-betaIT, an excitatory scorpion beta-toxin without a 'hot spot' at the relevant position
Biochem.Biophys.Res.Commun., 349, 2006
|
|
2DDY
 
 | | Solution Structure of Matrilysin (MMP-7) Complexed to Constraint Conformational Sulfonamide Inhibitor | | Descriptor: | (1R)-N,6-DIHYDROXY-7-METHOXY-2-[(4-METHOXYPHENYL)SULFONYL]-1,2,3,4-TETRAHYDROISOQUINOLINE-1-CARBOXAMIDE, CALCIUM ION, Matrilysin, ... | | Authors: | Zheng, X.H, Ou, L. | | Deposit date: | 2006-02-06 | | Release date: | 2007-02-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Matrilysin (MMP-7) Complexed to Constraint Conformational Sulfonamide Inhibitor
to be published
|
|
2EA2
 
 | | h-MetAP2 complexed with A773812 | | Descriptor: | 3-ETHYL-6-{[(4-FLUOROPHENYL)SULFONYL]AMINO}-2-METHYLBENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2007-01-30 | | Release date: | 2008-02-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lead optimization of methionine aminopeptidase-2 (MetAP2) inhibitors containing sulfonamides of 5,6-disubstituted anthranilic acids
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2EA4
 
 | | h-MetAP2 complexed with A797859 | | Descriptor: | 2-(2-AMINOETHOXY)-3-ETHYL-6-{[(4-FLUOROPHENYL)SULFONYL]AMINO}BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2007-01-30 | | Release date: | 2008-02-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Lead optimization of methionine aminopeptidase-2 (MetAP2) inhibitors containing sulfonamides of 5,6-disubstituted anthranilic acids
Bioorg.Med.Chem.Lett., 17, 2007
|
|
