6K3J
 
 | | Solution structure of APOBEC3G-CD2 with ssDNA, Product A | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*UP*(IUR)P*AP*AP*TP*T)-3'), DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Cao, C, Yan, X, Lan, W, Wang, C. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Investigations on the Interactions between Cytidine Deaminase Human APOBEC3G and DNA.
Chem Asian J, 14, 2019
|
|
8KDB
 
 | | Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in dimeric form | | Descriptor: | MAGNESIUM ION, Phosphoprotein, RNA-directed RNA polymerase L, ... | | Authors: | Xie, J, Wang, L, Zhai, G, Wu, D, Lin, Z, Wang, M, Yan, X, Gao, L, Huang, X, Fearns, R, Chen, S. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for dimerization of a paramyxovirus polymerase complex.
Nat Commun, 15, 2024
|
|
8KDC
 
 | | Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in monomeric form | | Descriptor: | MAGNESIUM ION, Phosphoprotein, RNA-directed RNA polymerase L, ... | | Authors: | Xie, J, Wang, L, Zhai, G, Wu, D, Lin, Z, Wang, M, Yan, X, Gao, L, Huang, X, Fearns, R, Chen, S. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for dimerization of a paramyxovirus polymerase complex.
Nat Commun, 15, 2024
|
|
2X7I
 
 | | Crystal structure of mevalonate kinase from methicillin-resistant Staphylococcus aureus MRSA252 | | Descriptor: | CHLORIDE ION, CITRIC ACID, MEVALONATE KINASE | | Authors: | Oke, M, Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-27 | | Release date: | 2010-07-21 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
6DXK
 
 | | Glucocorticoid Receptor in complex with Compound 11 | | Descriptor: | (8S,11R,13S,14S,17S)-11-[4-(dimethylamino)phenyl]-17-(3,3-dimethylbut-1-yn-1-yl)-17-hydroxy-13-methyl-1,2,6,7,8,11,12,13,14,15,16,17-dodecahydro-3H-cyclopenta[a]phenanthren-3-one (non-preferred name), Glucocorticoid receptor | | Authors: | Rew, Y, Du, X, Eksterowicz, J, Zhou, H, Jahchan, N, Zhu, L, Yan, X, Kawai, H, McGee, L.R, Medina, J.C, Huang, T, Chen, C, Zavorotinskaya, T, Sutimantanapi, D, Waszczuk, J, Jackson, E, Huang, E, Ye, Q, Fantin, V.R, Daqing, S. | | Deposit date: | 2018-06-29 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of a Potent and Selective Steroidal Glucocorticoid Receptor Antagonist (ORIC-101).
J. Med. Chem., 61, 2018
|
|
1M4X
 
 | | PBCV-1 virus capsid, quasi-atomic model | | Descriptor: | PBCV-1 virus capsid | | Authors: | Nandhagopal, N, Simpson, A.A, Gurnon, J.R, Yan, X, Baker, T.S, Graves, M.V, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2002-07-05 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | The Structure and Evolution of the Major Capsid Protein of a Large,
Lipid containing, DNA virus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2JOO
 
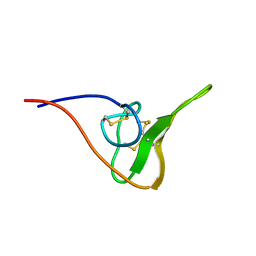 | | The NMR Solution Structure of Recombinant RGD-hirudin | | Descriptor: | Hirudin variant-1 | | Authors: | Song, X, Mo, W, Liu, X, Yan, X, Song, H, Dai, L. | | Deposit date: | 2007-03-14 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of recombinant RGD-hirudin
Biochem.Biophys.Res.Commun., 360, 2007
|
|
6OVH
 
 | | Cryo-EM structure of Bimetallic dodecameric cage design 3 (BMC3) from cytochrome cb562 | | Descriptor: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | Authors: | Golub, E, Subramanian, R.H, Yan, X, Alberstein, R.G, Tezcan, F.A. | | Deposit date: | 2019-05-07 | | Release date: | 2020-01-29 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
3SV1
 
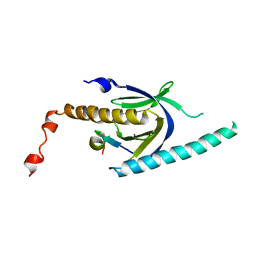 | | Crystal structure of APP peptide bound rat Mint2 PARM | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 2, Amyloid beta A4 protein | | Authors: | Shen, Y, Long, J, Yan, X, Xie, X. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Open-closed motion of Mint2 regulates APP metabolism
J Mol Cell Biol, 5, 2013
|
|
3SUZ
 
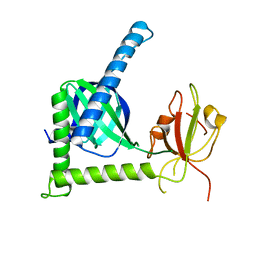 | | Crystal structure of Rat Mint2 PPC | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 2 | | Authors: | Shen, Y, Long, J, Yan, X, Xie, X. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Open-closed motion of Mint2 regulates APP metabolism
J Mol Cell Biol, 5, 2013
|
|
6HBA
 
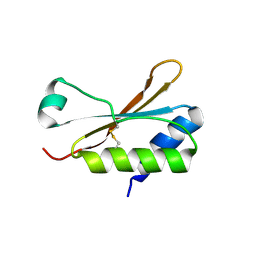 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942), thiol-oxidized form | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
3OKG
 
 | | Crystal structure of HsdS subunit from Thermoanaerobacter tengcongensis | | Descriptor: | Restriction endonuclease S subunits, SULFATE ION | | Authors: | Liang, D, Gao, P, Tang, Q, An, X, Yan, X. | | Deposit date: | 2010-08-24 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of HsdS subunit from Thermoanaerobacter tengcongensis sheds lights on mechanism of dynamic opening and closing of type I methyltransferase
Plos One, 6, 2011
|
|
8EO9
 
 | | The solution structure of abxF, an enzyme catalyzing the formation of chiral spiroketal of an antibiotics, (-)-ABX | | Descriptor: | Glyoxalase | | Authors: | Jia, X, Yan, X, Mobli, M, Qu, X. | | Deposit date: | 2022-10-02 | | Release date: | 2024-04-03 | | Method: | SOLUTION NMR | | Cite: | The solution structure of abxF, an enzyme catalyzing the formation of chiral spiroketal of an antibiotics, (-)-ABX.
To Be Published
|
|
8EPY
 
 | | The solution structure of abxF in complex with its product (-)-ABX, an enzyme catalyzing the formation of the chiral spiroketal of an anthrabenzoxocinone antibiotic, (-)-ABX | | Descriptor: | (6R,16R)-3,11,13,15-tetrahydroxy-1,6,9,9-tetramethyl-6,7,9,16-tetrahydro-14H-6,16-epoxyanthra[2,3-e]benzo[b]oxocin-14-one, Glyoxalase | | Authors: | Jia, X, Yan, X, Qu, X, Mobli, M. | | Deposit date: | 2022-10-06 | | Release date: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The solution structure of abxF, an enzyme catalyzing the formation of chiral spiroketal of an antibiotics, (-)-ABX.
To Be Published
|
|
7EPT
 
 | | Structural basis for the tethered peptide activation of adhesion GPCRs | | Descriptor: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ping, Y.-Q, Xiao, P, Yang, F, Zhao, R.-J, Guo, S.-C, Yan, X, Wu, X, Liebscher, I, Xu, H.E, Sun, J.-P. | | Deposit date: | 2021-04-27 | | Release date: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the tethered peptide activation of adhesion GPCRs.
Nature, 604, 2022
|
|
7CX3
 
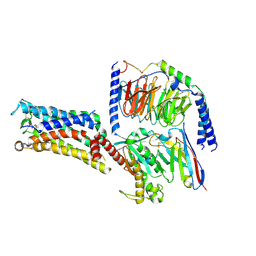 | | Cryo-EM structure of the Taprenepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-pyrazol-1-ylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX2
 
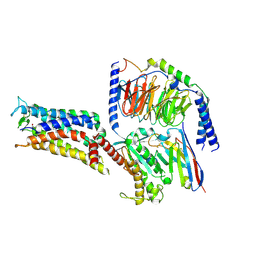 | | Cryo-EM structure of the PGE2-bound EP2-Gs complex | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX4
 
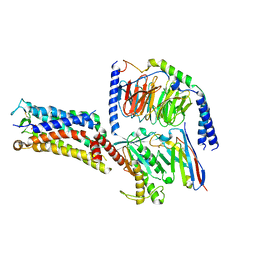 | | Cryo-EM structure of the Evatanepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-~{tert}-butylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
6LYN
 
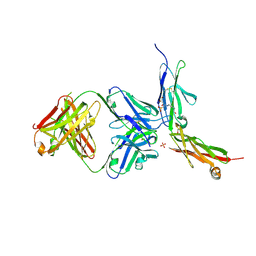 | | CD146 D4-D5/AA98 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AA98 Fab heavy chain, ... | | Authors: | Chen, X, Yan, X. | | Deposit date: | 2020-02-14 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.776 Å) | | Cite: | Structure basis for AA98 inhibition on the activation of endothelial cells mediated by CD146.
Iscience, 24, 2021
|
|
7CK9
 
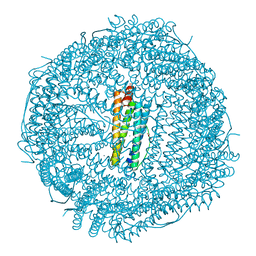 | | Crystal structure of Doxorubicin loaded human ferritin heavy chain | | Descriptor: | CHLORIDE ION, Ferritin heavy chain, GLYCEROL, ... | | Authors: | Chen, X, Jiang, B, Yan, X, Fan, K. | | Deposit date: | 2020-07-16 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | A natural drug entry channel in the ferritin nanocage.
Nano Today, 35, 2020
|
|
7CK8
 
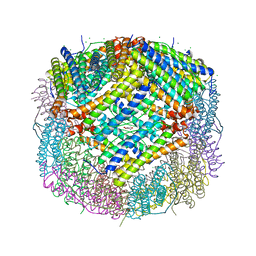 | | Crystal structure of human ferritin heavy chain mutant C90S/C102S/C130S | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin heavy chain, ... | | Authors: | Chen, X, Jiang, B, Yan, X, Fan, K. | | Deposit date: | 2020-07-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A natural drug entry channel in the ferritin nanocage.
Nano Today, 35, 2020
|
|
7XYT
 
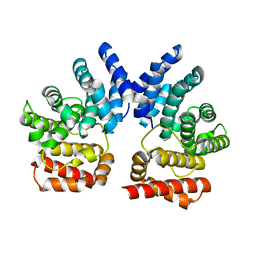 | |
7YNX
 
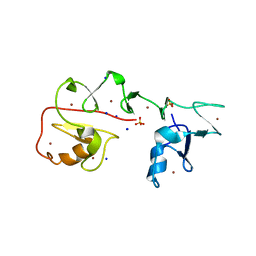 | | Crystal structure of Pirh2 bound to poly-Ala peptide | | Descriptor: | RING finger and CHY zinc finger domain-containing protein 1, SODIUM ION, SULFATE ION, ... | | Authors: | Dong, C, Yan, X, Li, Y. | | Deposit date: | 2022-08-01 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of an Ala-rich C-degron by the E3 ligase Pirh2.
Nat Commun, 14, 2023
|
|
6CLW
 
 | | Crystal structure of TnmH | | Descriptor: | O-methyltransferase | | Authors: | Chang, C.Y, Annaval, T, Adhikari, A, Yan, X, Shen, B. | | Deposit date: | 2018-03-02 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Characterization of TnmH as anO-Methyltransferase Revealing Insights into Tiancimycin Biosynthesis and Enabling a Biocatalytic Strategy To Prepare Antibody-Tiancimycin Conjugates.
J.Med.Chem., 63, 2020
|
|
6CLX
 
 | | Crystal structure of TnmH in complex with SAM | | Descriptor: | O-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Chang, C.Y, Annaval, T, Adhikari, A, Yan, X, Shen, B. | | Deposit date: | 2018-03-02 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Characterization of TnmH as anO-Methyltransferase Revealing Insights into Tiancimycin Biosynthesis and Enabling a Biocatalytic Strategy To Prepare Antibody-Tiancimycin Conjugates.
J.Med.Chem., 63, 2020
|
|
