6BUH
 
 | |
6CF6
 
 | | RNF146 TBM-Tankyrase ARC2-3 complex | | Descriptor: | RNF146, Tankyrase-1 | | Authors: | Da Rosa, P.A, Xu, W. | | Deposit date: | 2018-02-13 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for tankyrase-RNF146 interaction reveals noncanonical tankyrase-binding motifs.
Protein Sci., 27, 2018
|
|
7XMN
 
 | | Structure of SARS-CoV-2 ORF8 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltodextrin-binding protein, ... | | Authors: | Chen, X, Xu, W. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycosylated, Lipid-Binding, CDR-Like Domains of SARS-CoV-2 ORF8 Indicate Unique Sites of Immune Regulation.
Microbiol Spectr, 11, 2023
|
|
6KHX
 
 | | Crystal structure of Prx from Akkermansia muciniphila | | Descriptor: | CALCIUM ION, Peroxiredoxin | | Authors: | Li, M, Wang, J, Xu, W, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of Akkermansia muciniphila peroxiredoxin reveals a novel regulatory mechanism of typical 2-Cys Prxs by a distinct loop.
Febs Lett., 594, 2020
|
|
6LND
 
 | | Crystal structure of transposition protein TniQ | | Descriptor: | ZINC ION, transposition protein TniQ | | Authors: | Wang, B, Xu, W, Yang, H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex.
Cell Res., 30, 2020
|
|
6LNB
 
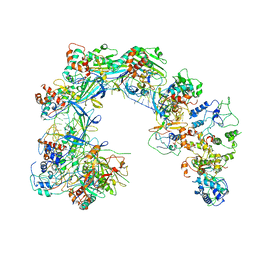 | | CryoEM structure of Cascade-TniQ-dsDNA complex | | Descriptor: | CRISPR RNA (60-MER), CRISPR-associated protein Cas6, CRISPR-associated protein Cas7, ... | | Authors: | Wang, B, Xu, W, Yang, H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex.
Cell Res., 30, 2020
|
|
6LNC
 
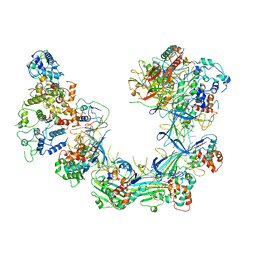 | | CryoEM structure of Cascade-TniQ complex | | Descriptor: | CRISPR RNA (60-MER), CRISPR-associated protein Cas6, CRISPR-associated protein Cas7, ... | | Authors: | Wang, B, Xu, W, Yang, H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex.
Cell Res., 30, 2020
|
|
2QBR
 
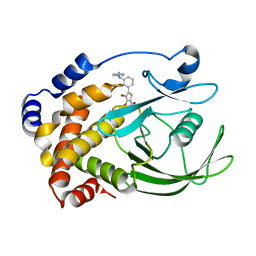 | | Crystal structure of ptp1b-inhibitor complex | | Descriptor: | 5-[3-(BENZYLAMINO)PHENYL]-4-BROMO-3-(CARBOXYMETHOXY)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W. | | Deposit date: | 2007-06-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site.
J.Med.Chem., 50, 2007
|
|
2QBQ
 
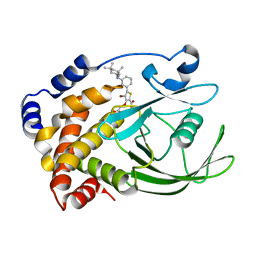 | | Crystal structure of ptp1b-inhibitor complex | | Descriptor: | 4-BROMO-3-(CARBOXYMETHOXY)-5-{3-[(3,3,5,5-TETRAMETHYLCYCLOHEXYL)AMINO]PHENYL}THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W. | | Deposit date: | 2007-06-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site.
J.Med.Chem., 50, 2007
|
|
2QBP
 
 | | Crystal structure of ptp1b-inhibitor complex | | Descriptor: | 5-(3-{[1-(BENZYLSULFONYL)PIPERIDIN-4-YL]AMINO}PHENYL)-4-BROMO-3-(CARBOXYMETHOXY)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W. | | Deposit date: | 2007-06-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site.
J.Med.Chem., 50, 2007
|
|
2QBS
 
 | | Crystal structure of ptp1b-inhibitor complex | | Descriptor: | 4-BROMO-3-(CARBOXYMETHOXY)-5-[3-(CYCLOHEXYLAMINO)PHENYL]THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W. | | Deposit date: | 2007-06-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site.
J.Med.Chem., 50, 2007
|
|
4TQS
 
 | |
4TQR
 
 | |
7XP5
 
 | | Cryo-EM structure of a class T GPCR in ligand-free state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP6
 
 | | Cryo-EM structure of a class T GPCR in active state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP4
 
 | | Cryo-EM structure of a class T GPCR in apo state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
6PUG
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1-2`OH-Ethyl-5-OP-U | | Descriptor: | 6-[(2-hydroxyethyl)amino]-5-[(E)-(2-oxopropylidene)amino]pyrimidine-2,4(1H,3H)-dione, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
1QB1
 
 | |
1QBN
 
 | |
1QB6
 
 | | BOVINE TRYPSIN 3,3'-[3,5-DIFLUORO-4-METHYL-2, 6-PYRIDINEDIYLBIS(OXY)]BIS(BENZENECARBOXIMIDAMIDE) (ZK-805623) COMPLEX | | Descriptor: | 3,3'-[3,5-DIFLUORO-4-METHYL-2,6-PYRIDYLENEBIS(OXY)]-BIS(BENZENECARBOXIMIDAMIDE), CALCIUM ION, POTASSIUM ION, ... | | Authors: | Whitlow, M. | | Deposit date: | 1999-04-29 | | Release date: | 2000-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic analysis of potent and selective factor Xa inhibitors complexed to bovine trypsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4AW5
 
 | | Complex of the EphB4 kinase domain with an oxindole inhibitor | | Descriptor: | (3Z)-5-[(1-ethylpiperidin-4-yl)amino]-3-[(5-methoxy-1H-benzimidazol-2-yl)(phenyl)methylidene]-1,3-dihydro-2H-indol-2-one, EPHRIN TYPE-B RECEPTOR 4 | | Authors: | Till, J.H, Stout, T.J. | | Deposit date: | 2012-05-31 | | Release date: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The Design, Synthesis, and Biological Evaluation of Potent Receptor Tyrosine Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6NTS
 
 | | Protein Phosphatase 2A (Aalpha-B56alpha-Calpha) holoenzyme in complex with a Small Molecule Activator of PP2A (SMAP) | | Descriptor: | MANGANESE (II) ION, N-[(1R,2R,3S)-2-hydroxy-3-(10H-phenoxazin-10-yl)cyclohexyl]-4-(trifluoromethoxy)benzene-1-sulfonamide, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2019-01-30 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Selective PP2A Enhancement through Biased Heterotrimer Stabilization.
Cell, 181, 2020
|
|
6PUK
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1-JYM72 | | Descriptor: | 1,2-dideoxy-1-{2,6-dioxo-5-[(1E)-3-oxobut-1-en-1-yl]-1,2,3,6-tetrahydropyrimidin-4-yl}-D-ribo-hexitol, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Awad, W, Keller, A.N, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
6PUF
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1-5'D-5-OP-RU | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
6PUM
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1-2'D-5-OP-RU | | Descriptor: | 1,2-dideoxy-1-({2,6-dioxo-5-[(E)-(2-oxopropylidene)amino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-erythro-pentitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Awad, W, Keller, A.N, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
