4U0L
 
 | |
4U0N
 
 | | Structure of the Vibrio cholerae di-nucleotide cyclase (DncV) deletion mutant D-loop | | Descriptor: | Cyclic AMP-GMP synthase, MAGNESIUM ION, N-[4-({[(6S)-2-amino-5-methyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-gamma-glutamyl-L-glutamic acid | | Authors: | Xiang, Y, Zhu, D.Y. | | Deposit date: | 2014-07-12 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural Biochemistry of a Vibrio cholerae Dinucleotide Cyclase Reveals Cyclase Activity Regulation by Folates.
Mol.Cell, 55, 2014
|
|
1T7B
 
 | | Crystal structure of mutant Lys8Gln of scorpion alpha-like neurotoxin BmK M1 from Buthus martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Xiang, Y, Guan, R.J, He, X.L, Wang, C.G, Wang, M, Zhang, Y, Sundberg, E.J, Wang, D.C. | | Deposit date: | 2004-05-09 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Mechanism Governing Cis and Trans Isomeric States and an Intramolecular Switch for Cis/Trans Isomerization of a Non-proline Peptide Bond Observed in Crystal Structures of Scorpion Toxins
J.Mol.Biol., 341, 2004
|
|
1T7A
 
 | | Crystal structure of mutant Lys8Asp of scorpion alpha-like neurotoxin BmK M1 from Buthus martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Xiang, Y, Guan, R.J, He, X.L, Wang, C.G, Wang, M, Zhang, Y, Sundberg, E.J, Wang, D.C. | | Deposit date: | 2004-05-08 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Mechanism Governing Cis and Trans Isomeric States and an Intramolecular Switch for Cis/Trans Isomerization of a Non-proline Peptide Bond Observed in Crystal Structures of Scorpion Toxins
J.Mol.Biol., 341, 2004
|
|
1T7E
 
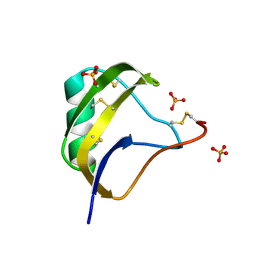 | | Crystal structure of mutant Pro9Ser of scorpion alpha-like neurotoxin BmK M1 from Buthus martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I, PHOSPHATE ION | | Authors: | Xiang, Y, Guan, R.J, He, X.L, Wang, C.G, Wang, M, Zhang, Y, Sundberg, E.J, Wang, D.C. | | Deposit date: | 2004-05-09 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Mechanism Governing Cis and Trans Isomeric States and an Intramolecular Switch for Cis/Trans Isomerization of a Non-proline Peptide Bond Observed in Crystal Structures of Scorpion Toxins
J.Mol.Biol., 341, 2004
|
|
8J1V
 
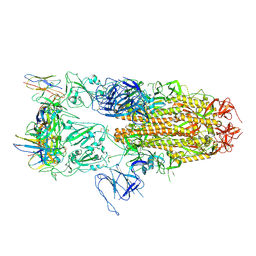 | |
8J1T
 
 | |
8IXJ
 
 | |
8IXL
 
 | | top segment of the bacteriophage M13 mini variant | | Descriptor: | Capsid protein G8P, Tail virion protein G7P, Tail virion protein G9P | | Authors: | Xiang, Y, Jia, Q. | | Deposit date: | 2023-04-01 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of a bacteriophage M13 mini variant.
Nat Commun, 14, 2023
|
|
8IXK
 
 | | bottom segment of the bacteriophage M13 mini variant | | Descriptor: | Attachment protein G3P, Capsid protein G8P, Head virion protein G6P | | Authors: | Xiang, Y, Jia, Q. | | Deposit date: | 2023-04-01 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of a bacteriophage M13 mini variant.
Nat Commun, 14, 2023
|
|
8JWT
 
 | |
7Y42
 
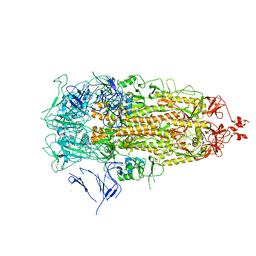 | |
6KW1
 
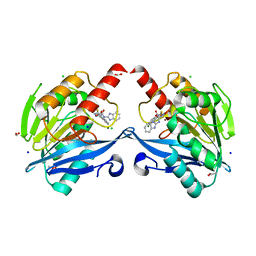 | | The structure of the metallo-beta-lactamase VIM-2 in complex with a triazolylthioacetamide 1b | | Descriptor: | 2-[3-[2-(1H-benzimidazol-2-ylamino)-2-oxidanylidene-ethyl]sulfanyl-1H-1,2,4-triazol-5-yl]benzoic acid, ACETATE ION, Beta-lactamase class B VIM-2, ... | | Authors: | Yang, K.W, Xiang, Y. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.775212 Å) | | Cite: | Kinetic, Thermodynamic, and Crystallographic Studies of 2-Triazolylthioacetamides as Verona Integron-Encoded Metallo-beta-Lactamase 2 (VIM-2) Inhibitor.
Biomolecules, 10, 2020
|
|
5Y66
 
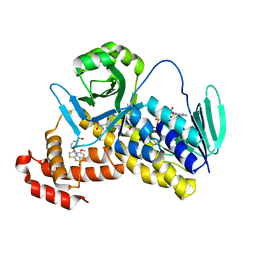 | | Crystal structure of Pseudomonas fluorescens Kynurenine 3-monooxygenase in complex with L-KYN and Ro61-8048 | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 3,4-dimethoxy-N-[4-(3-nitrophenyl)-1,3-thiazol-2-yl]benzenesulfonamide, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xiang, Y, Gao, J.J, Zhu, D.Y. | | Deposit date: | 2017-08-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Biochemistry and structural studies of kynurenine 3-monooxygenase reveal allosteric inhibition by Ro 61-8048
FASEB J., 32, 2018
|
|
5Y7A
 
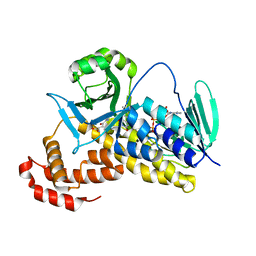 | |
5Y77
 
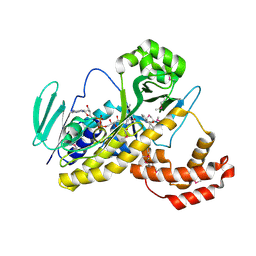 | |
7WCD
 
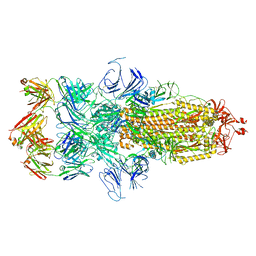 | | Cryo EM structure of SARS-CoV-2 spike in complex with TAU-2212 mAbs in conformation 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, Light chain, ... | | Authors: | Xiang, Y, Ma, B, Li, R. | | Deposit date: | 2021-12-19 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational flexibility in neutralization of SARS-CoV-2 by naturally elicited anti-SARS-CoV-2 antibodies.
Commun Biol, 5, 2022
|
|
7WC0
 
 | |
7WBZ
 
 | | Crystal structure of the SARS-Cov-2 RBD in complex with Fab 2303 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2303 heavy chain, 2303 light chain, ... | | Authors: | Xiang, Y, Ma, B. | | Deposit date: | 2021-12-17 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Conformational flexibility in neutralization of SARS-CoV-2 by naturally elicited anti-SARS-CoV-2 antibodies.
Commun Biol, 5, 2022
|
|
1KV0
 
 | | Cis/trans Isomerization of Non-prolyl Peptide Bond Observed in Crystal Structure of an Scorpion Toxin | | Descriptor: | Alpha-like toxin BmK-M7 | | Authors: | Guan, R.J, He, X.L, Wang, M, Xiang, Y, Wang, D.C. | | Deposit date: | 2002-01-23 | | Release date: | 2003-09-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural mechanism governing cis and trans isomeric states and an intramolecular switch for cis/trans isomerization of a non-proline peptide bond observed in crystal structures of scorpion toxins.
J.Mol.Biol., 341, 2004
|
|
4W60
 
 | | The structure of Vaccina virus H7 protein displays A Novel Phosphoinositide binding fold required for membrane biogenesis | | Descriptor: | Late protein H7 | | Authors: | Kolli, S, Meng, X, Wu, X, Shengjuler, D, Cameron, C.E, Xiang, Y, Deng, J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function analysis of vaccinia virus h7 protein reveals a novel phosphoinositide binding fold essential for poxvirus replication.
J.Virol., 89, 2015
|
|
1P9Z
 
 | | The Solution Structure of Antifungal Peptide Distinct With a Five-disulfide Motif from Eucommia ulmoides Oliver | | Descriptor: | Eucommia Antifungal peptide 2 | | Authors: | Huang, R.H, Xiang, Y, Tu, G.Z, Zhang, Y, Wang, D.C. | | Deposit date: | 2003-05-13 | | Release date: | 2004-05-25 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Eucommia Antifungal Peptide: A Novel Structural Model Distinct with a Five-Disulfide Motif.
Biochemistry, 43, 2004
|
|
4EEE
 
 | |
2ISO
 
 | | Ternary complex of DNA Polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-difluoromethylene triphosphate | | Descriptor: | 2'-DEOXY-5'-O-[({[DIFLUORO(PHOSPHONO)METHYL](HYDROXY)PHOSPHORYL}OXY)(HYDROXY)PHOSPHORYL]GUANOSINE, 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3', ... | | Authors: | Sucato, C.A, Upton, T.G, Kashemirov, B.A, Martinek, V, Xiang, Y, Beard, W.A. | | Deposit date: | 2006-10-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modifying the beta,gamma Leaving-Group Bridging Oxygen Alters Nucleotide Incorporation Efficiency, Fidelity, and the Catalytic Mechanism of DNA Polymerase beta.
Biochemistry, 46, 2007
|
|
2ISP
 
 | | Ternary complex of DNA Polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-methylene triphosphate | | Descriptor: | 2'-DEOXY-5'-O-(HYDROXY{[HYDROXY(PHOSPHONOMETHYL)PHOSPHORYL]OXY}PHOSPHORYL)GUANOSINE, 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3', ... | | Authors: | Sucato, C.A, Upton, T.G, Kashemirov, B.A, Martinek, V, Xiang, Y, Beard, W.A. | | Deposit date: | 2006-10-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Modifying the beta,gamma Leaving-Group Bridging Oxygen Alters Nucleotide Incorporation Efficiency, Fidelity, and the Catalytic Mechanism of DNA Polymerase beta.
Biochemistry, 46, 2007
|
|
