1PJW
 
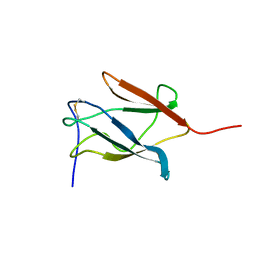 | | Solution Structure of the Domain III of the Japan Encephalitis Virus Envelope Protein | | Descriptor: | envelope protein | | Authors: | Wu, K.P, Wu, C.W, Tsao, Y.P, Lou, Y.C, Lin, C.W. | | Deposit date: | 2003-06-04 | | Release date: | 2003-11-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of a Flavivirus Recognized by Its Neutralizing Antibody: SOLUTION STRUCTURE OF THE DOMAIN III OF THE JAPANESE ENCEPHALITIS VIRUS ENVELOPE PROTEIN.
J.Biol.Chem., 278, 2003
|
|
1XHH
 
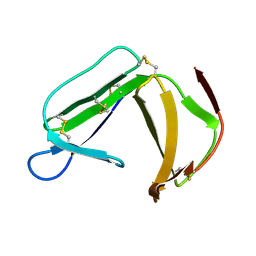 | | Solution Structure of porcine beta-microseminoprotein | | Descriptor: | beta-microseminoprotein | | Authors: | Wang, I, Lou, Y.C, Wu, K.P, Wu, S.H, Chang, W.C, Chen, C. | | Deposit date: | 2004-09-20 | | Release date: | 2005-03-20 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Novel solution structure of porcine beta-microseminoprotein
J.Mol.Biol., 346, 2005
|
|
9IUY
 
 | |
8JO2
 
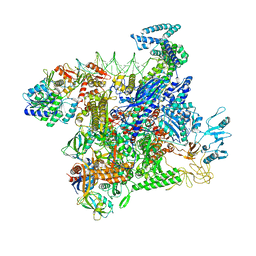 | | Structural basis of transcriptional activation by the OmpR/PhoB-family response regulator PmrA | | Descriptor: | DNA (65-MER), DNA-binding transcriptional regulator BasR, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lou, Y.-C, Huang, H.-Y, Chen, C, Wu, K.-P. | | Deposit date: | 2023-06-06 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis of transcriptional activation by the OmpR/PhoB-family response regulator PmrA.
Nucleic Acids Res., 51, 2023
|
|
7YAT
 
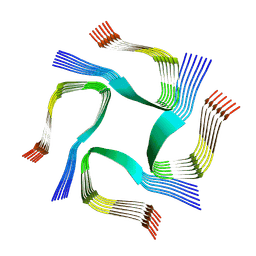 | | CryoEM tetra protofilament structure of the hamster prion 108-144 fibril | | Descriptor: | Major prion protein | | Authors: | Chen, E.H.-L, Kao, S.-W, Lee, C.-H, Huang, J.Y.C, Chen, R.P.-Y, Wu, K.-P. | | Deposit date: | 2022-06-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | 2.2 angstrom Cryo-EM Tetra-Protofilament Structure of the Hamster Prion 108-144 Fibril Reveals an Ordered Water Channel in the Center.
J.Am.Chem.Soc., 144, 2022
|
|
8J0A
 
 | | Robust design of effective allosteric activator UbV R4 for Rsp5 E3 ligase using the machine-learning tool ProteinMPNN | | Descriptor: | SULFATE ION, Ubiquitin variant R4 | | Authors: | Lin, Y.-F, Hsieh, Y.-J, Kao, H.-W, Ko, T.-P, Wu, K.-P. | | Deposit date: | 2023-04-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Robust Design of Effective Allosteric Activators for Rsp5 E3 Ligase Using the Machine Learning Tool ProteinMPNN.
Acs Synth Biol, 12, 2023
|
|
7YQM
 
 | |
7W85
 
 | |
5JG6
 
 | | APC11-Ubv shows role of noncovalent RING-Ubiquitin interactions in processive multiubiquitination and Ubiquitin chain elongation by APC/C | | Descriptor: | Anaphase-promoting complex subunit 11, Polyubiquitin-B, ZINC ION | | Authors: | Brown, N.G, Zhang, W, Yu, S, Miller, D.J, Sidhu, S.S, Schulman, B.A. | | Deposit date: | 2016-04-19 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0013 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
5L9U
 
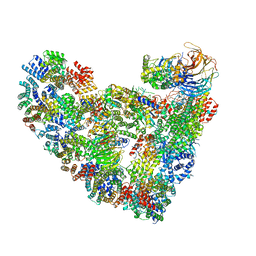 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with a cross linked Ubiquitin variant-substrate-UBE2C (UBCH10) complex representing key features of multiubiquitination | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
5L9T
 
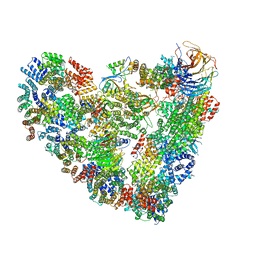 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with E2 UBE2S poised for polyubiquitination where UBE2S, APC2, and APC11 are modeled into low resolution density | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
7YQN
 
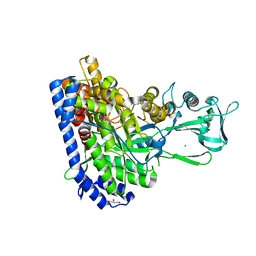 | | Crystal structure of Ecoli malate synthase G | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wu, K.-P, Lu, Y.-C, Ko, T.-P. | | Deposit date: | 2022-08-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cryo-EM reveals the structure and dynamics of a 723-residue malate synthase G.
J.Struct.Biol., 215, 2023
|
|
6JX7
 
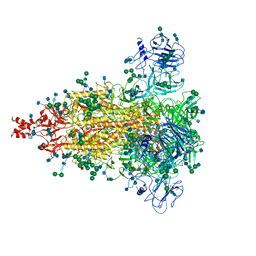 | | Cryo-EM structure of spike protein of feline infectious peritonitis virus strain UU4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Feline Infectious Peritonitis Virus Spike Protein, ... | | Authors: | Hsu, S.T.D, Yang, T.J, Ko, T.P, Draczkowski, P. | | Deposit date: | 2019-04-22 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM analysis of a feline coronavirus spike protein reveals a unique structure and camouflaging glycans.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
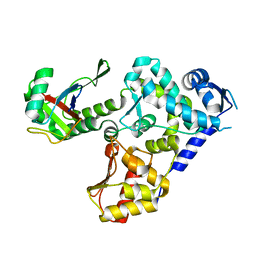
5HPS
 
 | |
5HPK
 
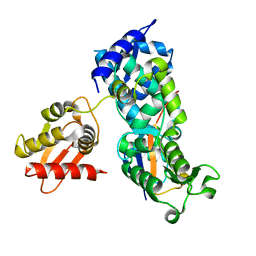 | | System-wide modulation of HECT E3 ligases with selective ubiquitin variant probes: NEDD4L and UbV NL.1 | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like, Ubiquitin variant NL.1 | | Authors: | Wu, K.-P, Mukherjee, M, Mercredi, P.Y, Schulman, B.A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.431 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
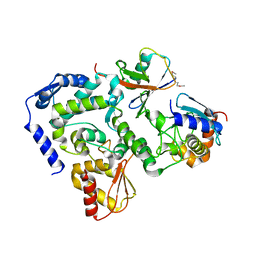
5HPT
 
 | |
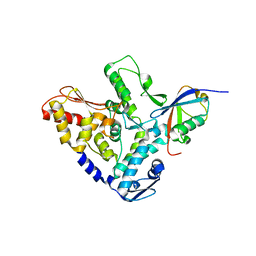
5HPL
 
 | |
7VPW
 
 | |
5V4K
 
 | | Crystal structure of NEDD4 LIR-fused human LC3B_2-119 | | Descriptor: | GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3B,Microtubule-associated proteins 1A/1B light chain 3B,Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Qiu, Y, Zheng, Y, Schulman, B. | | Deposit date: | 2017-03-09 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Insights into links between autophagy and the ubiquitin system from the structure of LC3B bound to the LIR motif from the E3 ligase NEDD4.
Protein Sci., 26, 2017
|
|
5C7M
 
 | | CRYSTAL STRUCTURE OF E3 LIGASE ITCH WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Wernimont, A, Zhang, W, Sidhu, S, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
5C7J
 
 | | CRYSTAL STRUCTURE OF NEDD4 WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
5IFR
 
 | | Structure of the stable UBE2D3-UbDha conjugate | | Descriptor: | GLYCEROL, Polyubiquitin-B, Ubiquitin-conjugating enzyme E2 D3 | | Authors: | Pruneda, J.N, Mulder, M.P.C, Witting, K, Ovaa, H, Komander, D. | | Deposit date: | 2016-02-26 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cascading activity-based probe sequentially targets E1-E2-E3 ubiquitin enzymes.
Nat.Chem.Biol., 12, 2016
|
|
