3KBH
 
 | | Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Wu, K, Li, W, Peng, G, Li, F. | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8KFK
 
 | |
3SCI
 
 | | Crystal structure of spike protein receptor-binding domain from a predicted SARS coronavirus human strain complexed with human receptor ACE2 | | Descriptor: | Angiotensin-converting enzyme 2, CHLORIDE ION, Spike glycoprotein, ... | | Authors: | Wu, K, Peng, G, Wilken, M, Geraghty, R, Li, F. | | Deposit date: | 2011-06-07 | | Release date: | 2012-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus.
J.Biol.Chem., 287, 2012
|
|
3SCK
 
 | | Crystal structure of spike protein receptor-binding domain from a predicted SARS coronavirus civet strain complexed with human-civet chimeric receptor ACE2 | | Descriptor: | Angiotensin-converting enzyme 2 chimera, CHLORIDE ION, Spike glycoprotein, ... | | Authors: | Wu, K, Peng, G, Wilken, M, Geraghty, R, Li, F. | | Deposit date: | 2011-06-07 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus.
J.Biol.Chem., 287, 2012
|
|
7WXE
 
 | |
3SCJ
 
 | | Crystal structure of spike protein receptor-binding domain from a predicted SARS coronavirus civet strain complexed with human receptor ACE2 | | Descriptor: | Angiotensin-converting enzyme 2, CHLORIDE ION, Spike glycoprotein, ... | | Authors: | Wu, K, Peng, G, Wilken, M, Geraghty, R, Li, F. | | Deposit date: | 2011-06-07 | | Release date: | 2012-02-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus.
J.Biol.Chem., 287, 2012
|
|
3SCL
 
 | | Crystal structure of spike protein receptor-binding domain from SARS coronavirus epidemic strain complexed with human-civet chimeric receptor ACE2 | | Descriptor: | Angiotensin-converting enzyme 2 chimera, CHLORIDE ION, Spike glycoprotein, ... | | Authors: | Wu, K, Peng, G, Wilken, M, Geraghty, R, Li, F. | | Deposit date: | 2011-06-07 | | Release date: | 2012-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus.
J.Biol.Chem., 287, 2012
|
|
2LRM
 
 | |
2LRV
 
 | |
8DSV
 
 | | The structure of NicA2 in complex with N-methylmyosmine | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-dependent oxidoreductase, ... | | Authors: | Wu, K, Dulchavsky, M, Li, J, Bardwell, J.C.A. | | Deposit date: | 2022-07-22 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme.
Nat.Chem.Biol., 19, 2023
|
|
8DQ8
 
 | | The structure of NicA2 variant F104L/A107T/S146I/G317D/H368R/L449V/N462S in complex with N-methylmyosmine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 1,2-ETHANEDIOL, Amine oxidase, ... | | Authors: | Wu, K, Dulchavsky, M, Li, J, Bardwell, J.C.A. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme.
Nat.Chem.Biol., 19, 2023
|
|
8DQ7
 
 | |
6X84
 
 | |
7TQR
 
 | |
7TLX
 
 | | Crystal Structure of cytochrome c from Pseudomonas putida S16 | | Descriptor: | C-type cytochrome, HEME C | | Authors: | Wu, K, Dulchavsky, M, Stull, F, Bardwell, J.C.A. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The enzyme pseudooxynicotine amine oxidase from Pseudomonas putida S16 is not an oxidase, but a dehydrogenase.
J.Biol.Chem., 298, 2022
|
|
7WPE
 
 | | SARS-CoV-2 Omicron Variant S Trimer complexed with two JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WRV
 
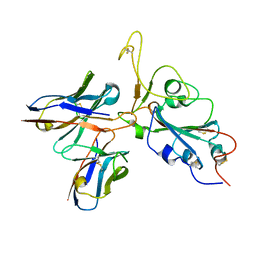 | | The interface of JMB2002 Fab binds to SARS-CoV-2 Omicron Variant S | | Descriptor: | JMB2002 Fab heavy chain, JMB2002 Fab light chain, Spike glycoprotein | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPB
 
 | | SARS-CoV-2 Omicron Variant RBD complexed with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPC
 
 | | The second RBD of SARS-CoV-2 Omicron Variant in complexed with RBD-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPA
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer complexed with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WP9
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer, all RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPF
 
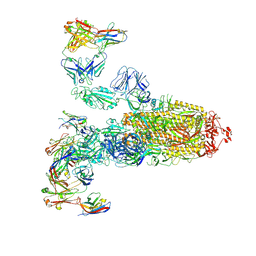 | | SARS-CoV-2 Omicron Variant S Trimer complexed with three JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPD
 
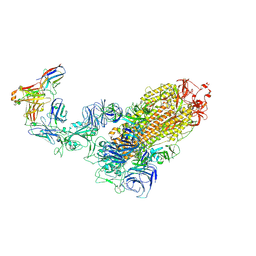 | | SARS-CoV-2 Omicron Variant S Trimer complexed with one JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
8JR9
 
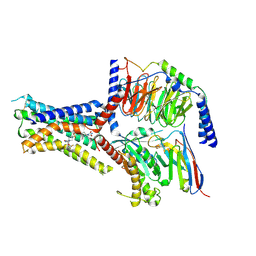 | | Small molecule agonist (PCO371) bound to human parathyroid hormone receptor type 1 (PTH1R) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha-1, ... | | Authors: | Zhao, L, He, Q, Yuan, Q, Gu, Y, He, X, Shan, H, Li, J, Wang, K, Li, Y, Hu, W, Wu, K, Shen, J, Xu, H.E. | | Deposit date: | 2023-06-16 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Conserved class B GPCR activation by a biased intracellular agonist.
Nature, 621, 2023
|
|
8WSS
 
 | | Cryo-EM structure of Melanin-Concentrating Hormone Receptor 1 with MCH | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, L, He, Q, Yuan, Q, Gu, Y, Shan, H, Hu, W, Wu, K, Xu, H.E, Zhang, Y, Wu, C. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Mechanisms of ligand recognition and activation of melanin-concentrating hormone receptors.
Cell Discov, 10, 2024
|
|
