8GW2
 
 | |
8H2R
 
 | |
1DXN
 
 | |
1MQX
 
 | | NMR Solution Structure of Type-B Lantibiotics Mersacidin in MeOH/H2O Mixture | | Descriptor: | LANTIBIOTIC MERSACIDIN | | Authors: | Hsu, S.-T, Breukink, E, Bierbaum, G, Sahl, H.-G, de Kruijff, B, Kaptein, R, van Nuland, N.A, Bonvin, A.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-03-11 | | Last modified: | 2018-08-08 | | Method: | SOLUTION NMR | | Cite: | NMR Study of Mersacidin and Lipid II Interaction in Dodecylphosphocholine Micelles. Conformational Changes are a Key to Antimicrobial Activity
J.Biol.Chem., 278, 2003
|
|
1MQZ
 
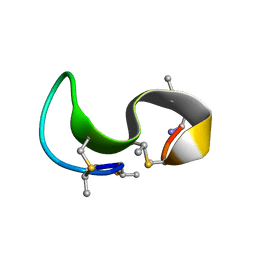 | | NMR solution structure of type-B lantibiotics mersacidin bound to lipid II in DPC micelles | | Descriptor: | LANTIBIOTIC MERSACIDIN | | Authors: | Hsu, S.-T, Breukink, E, Bierbaum, G, Sahl, H.-G, de Kruijff, B, Kaptein, R, van Nuland, N.A, Bonvin, A.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-03-11 | | Last modified: | 2018-08-08 | | Method: | SOLUTION NMR | | Cite: | NMR Study of Mersacidin and Lipid II Interaction in Dodecylphosphocholine Micelles. Conformational Changes are a Key to Antimicrobial Activity
J.Biol.Chem., 278, 2003
|
|
1MQY
 
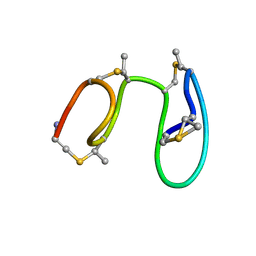 | | NMR solution structure of type-B lantibiotics mersacidin in DPC micelles | | Descriptor: | LANTIBIOTIC MERSACIDIN | | Authors: | Hsu, S.-T, Breukink, E, Bierbaum, G, Sahl, H.-G, de Kruijff, B, Kaptein, R, van Nuland, N.A, Bonvin, A.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-03-11 | | Last modified: | 2018-08-08 | | Method: | SOLUTION NMR | | Cite: | NMR Study of Mersacidin and Lipid II Interaction in Dodecylphosphocholine Micelles. Conformational Changes are a Key to Antimicrobial Activity
J.Biol.Chem., 278, 2003
|
|
1WCO
 
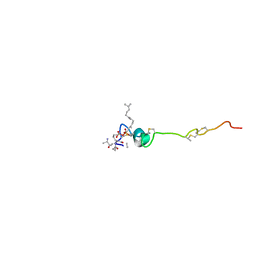 | | The solution structure of the nisin-lipid II complex | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, ALA-FGA-LYS-DAL-DAL PEPTIDE, ... | | Authors: | Hsu, S.-T.D, Breukink, E, Tischenko, E, Lutters, M.A.G, de Kruijff, B, Kaptein, R, Bonvin, A.M.J.J, van Nuland, N.A.J. | | Deposit date: | 2004-11-19 | | Release date: | 2005-03-07 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics.
Nat. Struct. Mol. Biol., 11, 2004
|
|
7Y6U
 
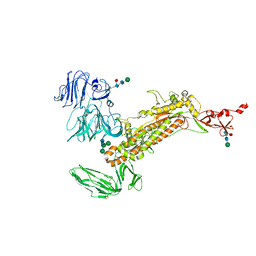 | | Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-close conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6V
 
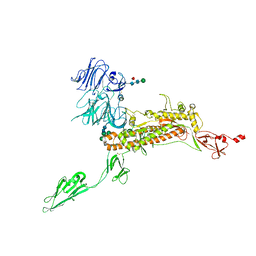 | | Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6T
 
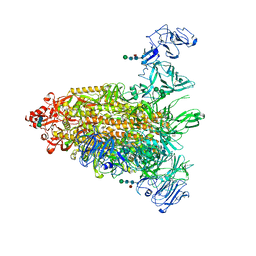 | | Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein one D0-down and two D0-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6S
 
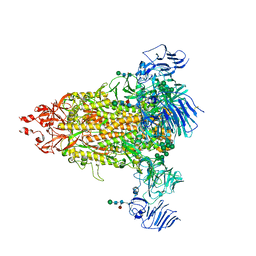 | | Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein with three D0-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7W73
 
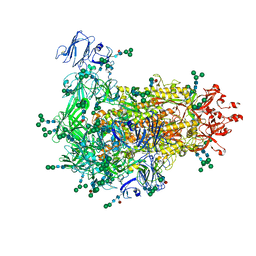 | | Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2021-12-03 | | Release date: | 2022-08-03 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7W6M
 
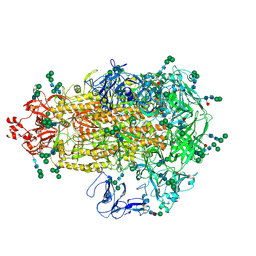 | | Cryo-EM map of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
2KQH
 
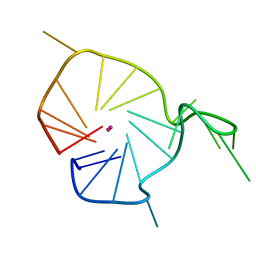 | | A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics | | Descriptor: | 5'-D(*CP*GP*GP*GP*CP*GP*GP*GP*CP*GP*CP*GP*AP*GP*GP*GP*AP*GP*GP*GP*T)-3', POTASSIUM ION | | Authors: | Hsu, S.-T.D, Varnai, P, Bugaut, A, Reszka, A.P, Neidle, S, Balasubramanian, S. | | Deposit date: | 2009-11-05 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics
J.Am.Chem.Soc., 131, 2009
|
|
2KQG
 
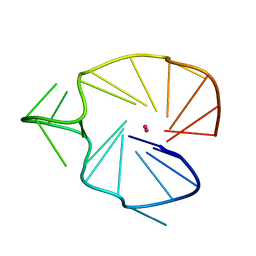 | | A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics | | Descriptor: | 5'-D(*CP*GP*GP*GP*CP*GP*GP*GP*CP*AP*CP*GP*AP*GP*GP*GP*AP*GP*GP*GP*T)-3', POTASSIUM ION | | Authors: | Hsu, S.-T.D, Varnai, P, Bugaut, A, Reszka, A.P, Neidle, S, Balasubramanian, S. | | Deposit date: | 2009-11-05 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics
J.Am.Chem.Soc., 131, 2009
|
|
6JX7
 
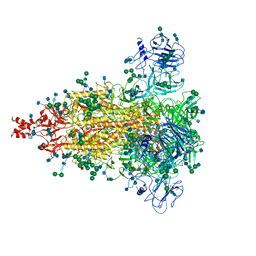 | | Cryo-EM structure of spike protein of feline infectious peritonitis virus strain UU4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Feline Infectious Peritonitis Virus Spike Protein, ... | | Authors: | Hsu, S.T.D, Yang, T.J, Ko, T.P, Draczkowski, P. | | Deposit date: | 2019-04-22 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM analysis of a feline coronavirus spike protein reveals a unique structure and camouflaging glycans.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7YMT
 
 | | Cryo-EM structure of MERS-CoV spike protein, Two RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (6.55 Å) | | Cite: | GlycoSHIELD: a versatile pipeline to assess glycan impact on protein structures
To Be Published
|
|
7YMW
 
 | | Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (6.05 Å) | | Cite: | GlycoSHIELD: a versatile pipeline to assess glycan impact on protein structures
To Be Published
|
|
7YMX
 
 | | Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.44 Å) | | Cite: | GlycoSHIELD: a versatile pipeline to assess glycan impact on protein structures
To Be Published
|
|
7YMV
 
 | | Cryo-EM structure of MERS-CoV spike protein, Two RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (6.74 Å) | | Cite: | GlycoSHIELD: a versatile pipeline to assess glycan impact on protein structures
To Be Published
|
|
7YN0
 
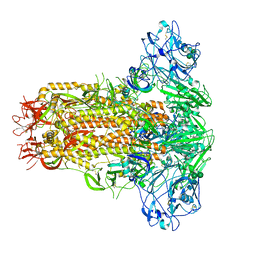 | | Cryo-EM structure of MERS-CoV spike protein, all RBD-down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | GlycoSHIELD: a versatile pipeline to assess glycan impact on protein structures
To Be Published
|
|
7YMY
 
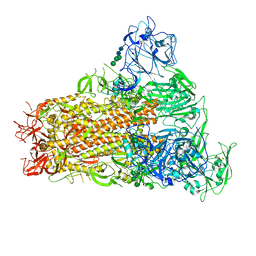 | | Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.96 Å) | | Cite: | GlycoSHIELD: a versatile pipeline to assess glycan impact on protein structures
To Be Published
|
|
7YMZ
 
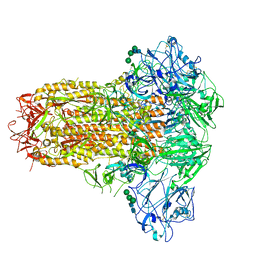 | | Cryo-EM structure of MERS-CoV spike protein, intermediate conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | GlycoSHIELD: a versatile pipeline to assess glycan impact on protein structures
To Be Published
|
|
8W68
 
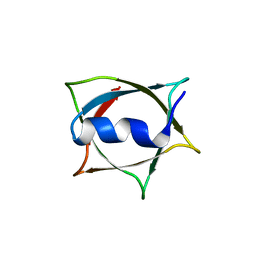 | | Crystal structure of Q9PR55 at pH 6.0 (use NMR model) | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-08-28 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8IWA
 
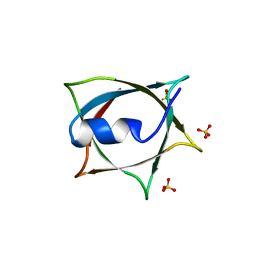 | | Crystal structure of Q9PR55 at pH 6.5 | | Descriptor: | SULFATE ION, Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
