4G84
 
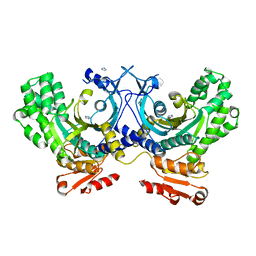 | | Crystal structure of human HisRS | | Descriptor: | CHLORIDE ION, Histidine--tRNA ligase, cytoplasmic, ... | | Authors: | Wei, Z, Wu, J, Zhou, J.J, Yang, X.-L, Zhang, M, Schimmel, P. | | Deposit date: | 2012-07-21 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Internally Deleted Human tRNA Synthetase Suggests Evolutionary Pressure for Repurposing.
Structure, 20, 2012
|
|
4G85
 
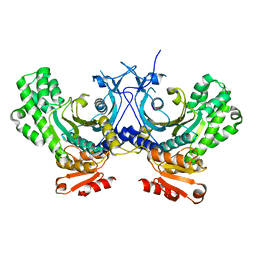 | | Crystal structure of human HisRS | | Descriptor: | Histidine-tRNA ligase, cytoplasmic | | Authors: | Wei, Z, Wu, J, Zhou, J.J, Yang, X.-L, Zhang, M, Schimmel, P. | | Deposit date: | 2012-07-21 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Internally Deleted Human tRNA Synthetase Suggests Evolutionary Pressure for Repurposing.
Structure, 20, 2012
|
|
4GMV
 
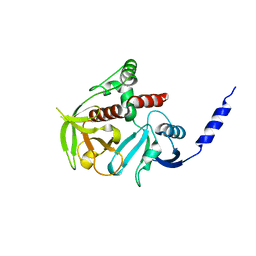 | |
4GN1
 
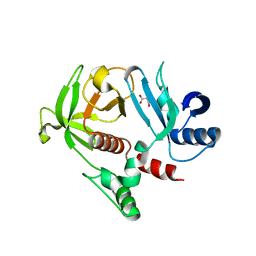 | | Crystal Structure of the RA and PH domains of Lamellipodin | | Descriptor: | MALONATE ION, Ras-associated and pleckstrin homology domains-containing protein 1 | | Authors: | Chang, Y.C.E, Wu, J. | | Deposit date: | 2012-08-16 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Lamellipodin implicates diverse functions in actin polymerization and Ras signaling.
Protein Cell, 4, 2013
|
|
4GXL
 
 | | The crystal structure of Galectin-8 C-CRD in complex with NDP52 | | Descriptor: | GLYCEROL, Galectin-8, Peptide from Calcium-binding and coiled-coil domain-containing protein 2 | | Authors: | Li, S, Wandel, M.P, Li, F, Liu, Z, He, C, Wu, J, Shi, Y, Randow, F. | | Deposit date: | 2012-09-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.023 Å) | | Cite: | Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy
Sci.Signal., 6, 2013
|
|
4HPT
 
 | | Crystal structure of the catalytic subunit of cAMP-dependent protein kinase displaying complete phosphoryl transfer of AMP-PNP onto a substrate peptide | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Bastidas, A.C, Steichen, J.M, Wu, J, Taylor, S.S. | | Deposit date: | 2012-10-24 | | Release date: | 2013-03-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Phosphoryl transfer by protein kinase a is captured in a crystal lattice.
J.Am.Chem.Soc., 135, 2013
|
|
4HPU
 
 | | Crystal structure of the catalytic subunit of cAMP-dependent protein kinase displaying partial phosphoryl transfer of AMP-PNP onto a substrate peptide | | Descriptor: | MAGNESIUM ION, MYRISTIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Bastidas, A.C, Steichen, J.M, Wu, J, Taylor, S.S. | | Deposit date: | 2012-10-24 | | Release date: | 2013-03-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Phosphoryl transfer by protein kinase a is captured in a crystal lattice.
J.Am.Chem.Soc., 135, 2013
|
|
4JVA
 
 | | Crystal Structure of RIIbeta(108-402) bound to HE33, a N6 di-propyl substituted cAMP analog | | Descriptor: | (2R,4aR,6R,7R,7aS)-6-[6-(dipropylamino)-9H-purin-9-yl]tetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinine-2,7-diol 2-oxide, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Brown, S.H.J, Cheng, C.Y, Saldanha, A.S, Wu, J, Cottam, H, Sankaran, B, Taylor, S.S. | | Deposit date: | 2013-03-25 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Implementing Fluorescence Anisotropy Screening and Crystallographic Analysis to Define PKA Isoform-Selective Activation by cAMP Analogs.
Acs Chem.Biol., 8, 2013
|
|
4JV4
 
 | | Crystal Structure of RIalpha(91-379) bound to HE33, a N6 di-propyl substituted cAMP analog | | Descriptor: | (2R,4aR,6R,7R,7aS)-6-[6-(dipropylamino)-9H-purin-9-yl]tetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinine-2,7-diol 2-oxide, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Brown, S.H.J, Cheng, C.Y, Saldanha, A.S, Wu, J, Cottam, H, Sankaran, B, Taylor, S.S. | | Deposit date: | 2013-03-25 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Implementing Fluorescence Anisotropy Screening and Crystallographic Analysis to Define PKA Isoform-Selective Activation by cAMP Analogs.
Acs Chem.Biol., 8, 2013
|
|
5Y59
 
 | | Crystal structure of Ku80 and Sir4 | | Descriptor: | ATP-dependent DNA helicase II subunit 2, SULFATE ION, Sir4p | | Authors: | Chen, H, Xue, J, Wu, J, Lei, M. | | Deposit date: | 2017-08-08 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structural Insights into Yeast Telomerase Recruitment to Telomeres
Cell, 172, 2018
|
|
5Y5A
 
 | | Crystal structure of Est1 and Cdc13 | | Descriptor: | KLLA0F20702p, KLLA0F20922p | | Authors: | Chen, H, Xue, J, Wu, J, Lei, M. | | Deposit date: | 2017-08-08 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural Insights into Yeast Telomerase Recruitment to Telomeres
Cell, 172, 2018
|
|
5Y58
 
 | | Crystal structure of Ku70/80 and TLC1 | | Descriptor: | ATP-dependent DNA helicase II subunit 1, ATP-dependent DNA helicase II subunit 2, TLC1 | | Authors: | Chen, H, Xue, J, Wu, J, Lei, M. | | Deposit date: | 2017-08-08 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into Yeast Telomerase Recruitment to Telomeres
Cell, 172, 2018
|
|
5WZG
 
 | | Structure of APUM23-GAAUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*AP*AP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
5WZJ
 
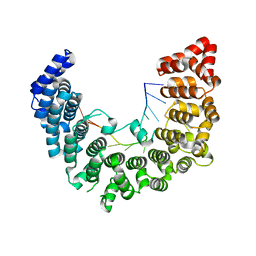 | | Structure of APUM23-GGAUUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*UP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
5WZH
 
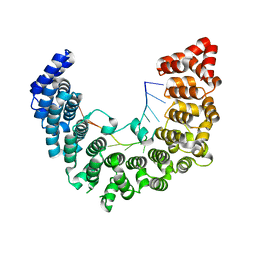 | | Structure of APUM23-GGAAUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*AP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
5WZI
 
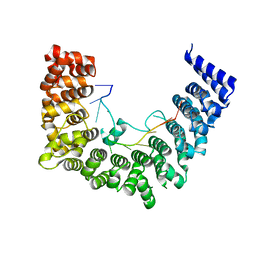 | | Structure of APUM23-GGAGUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*GP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
5WZK
 
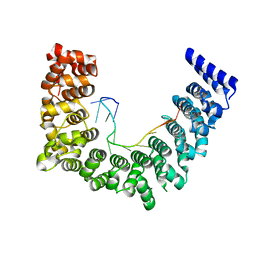 | | Structure of APUM23-deletion-of-insert-region-GGAAUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*AP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
7BU5
 
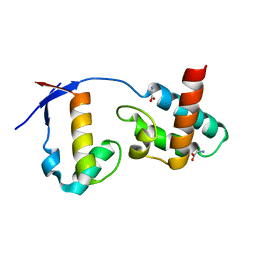 | | Crystal Structure of Human SLX4 and MUS81 | | Descriptor: | GLYCINE, MUS81 endonuclease homolog (Yeast), isoform CRA_b, ... | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2020-04-04 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SLX4 control MUS81 endonuclease function in telomere through an intermolecular SAP domain
To be published
|
|
7C7A
 
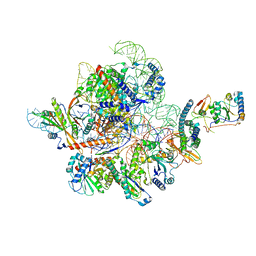 | | Cryo-EM structure of yeast Ribonuclease MRP with substrate ITS1 | | Descriptor: | Internal Transcribed Spacer 1, MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, ... | | Authors: | Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-05-24 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight into precursor ribosomal RNA processing by ribonuclease MRP.
Science, 369, 2020
|
|
7C79
 
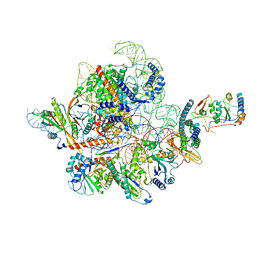 | | Cryo-EM structure of yeast Ribonuclease MRP | | Descriptor: | MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, Ribonuclease MRP RNA subunit NME1, ... | | Authors: | Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-05-24 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insight into precursor ribosomal RNA processing by ribonuclease MRP.
Science, 369, 2020
|
|
5Y1U
 
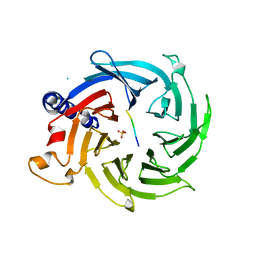 | | Crystal structure of RBBP4 bound to AEBP2 RRK motif | | Descriptor: | Histone-binding protein RBBP4, SULFATE ION, Zinc finger protein AEBP2 | | Authors: | Sun, A, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2017-07-21 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural and biochemical insights into human zinc finger protein AEBP2 reveals interactions with RBBP4
Protein Cell, 2017
|
|
5Y0U
 
 | | The solution structure of AEBP2 C2H2 zinc fingers | | Descriptor: | ZINC ION, Zinc finger protein AEBP2 | | Authors: | Sun, A, Shi, Y, Wu, J. | | Deposit date: | 2017-07-18 | | Release date: | 2018-08-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and biochemical insights into human zinc finger protein AEBP2 reveals interactions with RBBP4.
Protein Cell, 9, 2018
|
|
6AH3
 
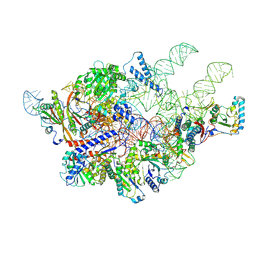 | | Cryo-EM structure of yeast Ribonuclease P with pre-tRNA substrate | | Descriptor: | MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
6AGB
 
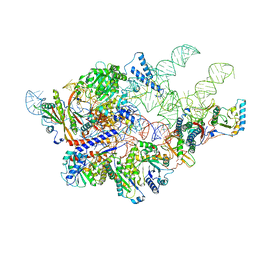 | | Cryo-EM structure of yeast Ribonuclease P | | Descriptor: | RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, Ribonuclease P protein subunit RPR2, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
6BYM
 
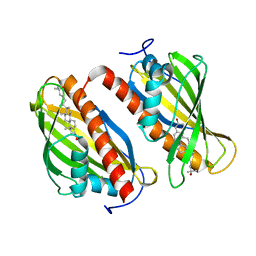 | | Crystal structure of the sterol-bound second StART domain of yeast Lam4 | | Descriptor: | 25-HYDROXYCHOLESTEROL, Sterol-binding protein | | Authors: | Jentsch, J.A, Kiburu, I.N, Wu, J, Pandey, K, Boudker, O, Menon, A.K. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of sterol binding and transport by a yeast StARkin domain.
J. Biol. Chem., 293, 2018
|
|
