1S8F
 
 | |
2QCC
 
 | |
2QCL
 
 | |
2QCM
 
 | |
2QCF
 
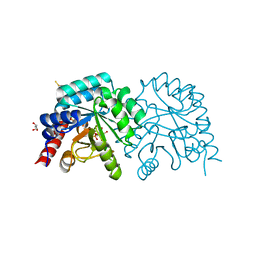 | |
2QCE
 
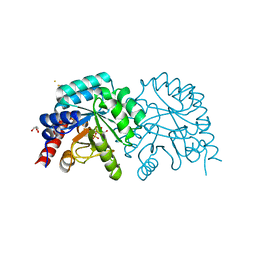 | | Crystal structure of the orotidine-5'-monophosphate decarboxylase domain of human UMP synthase bound to sulfate, glycerol, and chloride | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Wittmann, J, Rudolph, M. | | Deposit date: | 2007-06-19 | | Release date: | 2007-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structures of the human orotidine-5'-monophosphate decarboxylase support a covalent mechanism and provide a framework for drug design.
Structure, 16, 2008
|
|
2QCG
 
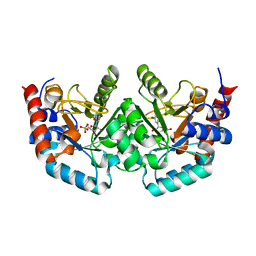 | |
2QCH
 
 | |
2QCN
 
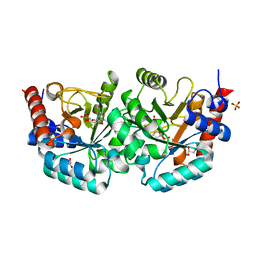 | |
2QCD
 
 | |
3LLL
 
 | | Crystal structure of mouse pacsin2 F-BAR domain | | Descriptor: | CALCIUM ION, CHLORIDE ION, Protein kinase C and casein kinase substrate in neurons protein 2, ... | | Authors: | Rudolph, M.G. | | Deposit date: | 2010-01-29 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A hinge in the distal end of the PACSIN 2 F-BAR domain may contribute to membrane-curvature sensing.
J.Mol.Biol., 400, 2010
|
|
2GXS
 
 | |
2GXU
 
 | |
2GXQ
 
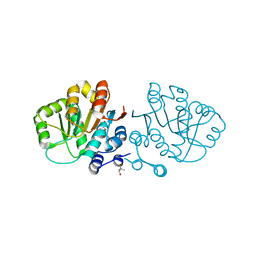 | | HERA N-terminal domain in complex with AMP, crystal form 1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, heat resistant RNA dependent ATPase | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2006-05-09 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure and Nucleotide Binding of the Thermus thermophilus RNA Helicase Hera N-terminal Domain.
J.Mol.Biol., 351, 2006
|
|
2AFT
 
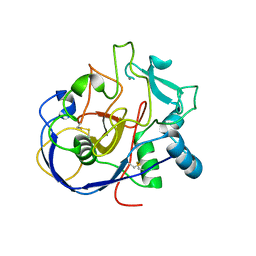 | | Formylglycine generating enzyme C336S mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Sulfatase modifying factor 1 | | Authors: | Roeser, D, Rudolph, M.G. | | Deposit date: | 2005-07-26 | | Release date: | 2005-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A general binding mechanism for all human sulfatases by the formylglycine-generating enzyme
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2AFY
 
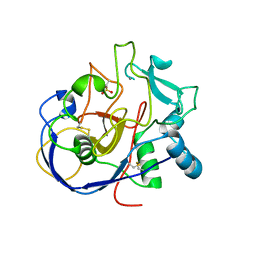 | | Formylglycine generating enzyme C341S mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Sulfatase modifying factor 1 | | Authors: | Roeser, D, Rudolph, M.G. | | Deposit date: | 2005-07-26 | | Release date: | 2005-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A general binding mechanism for all human sulfatases by the formylglycine-generating enzyme
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2AII
 
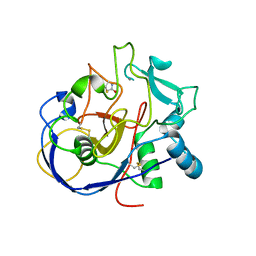 | | wild-type Formylglycine generating enzyme reacted with iodoacetamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETAMIDE, CALCIUM ION, ... | | Authors: | Roeser, D, Rudolph, M.G. | | Deposit date: | 2005-07-29 | | Release date: | 2005-12-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A general binding mechanism for all human sulfatases by the formylglycine-generating enzyme
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2AIK
 
 | |
2AIJ
 
 | |
