1F4K
 
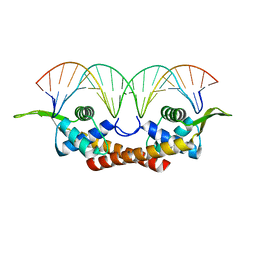 | | CRYSTAL STRUCTURE OF THE REPLICATION TERMINATOR PROTEIN/B-SITE DNA COMPLEX | | Descriptor: | 5'-D(*CP*TP*AP*TP*GP*AP*AP*CP*AP*TP*AP*AP*TP*GP*TP*TP*CP*AP*TP*AP*G)-3', 5'-D(*CP*TP*AP*TP*GP*AP*AP*CP*AP*TP*TP*AP*TP*GP*TP*TP*CP*AP*TP*AP*G)-3', REPLICATION TERMINATION PROTEIN | | Authors: | Wilce, J.A, Vivian, J.P, Hastings, A.F, Otting, G, Folmer, R.H.A, Duggin, I.G, Wake, R.G, Wilce, M.C.J. | | Deposit date: | 2000-06-08 | | Release date: | 2001-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the RTP-DNA complex and the mechanism of polar replication fork arrest
Nat.Struct.Biol., 8, 2001
|
|
3PQZ
 
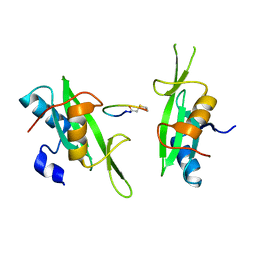 | | Grb7 SH2 with peptide | | Descriptor: | Growth factor receptor-bound protein 7, cyclic peptide | | Authors: | Wilce, J.A. | | Deposit date: | 2010-11-29 | | Release date: | 2011-07-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structural basis of binding by cyclic nonphosphorylated Peptide antagonists of grb7 implicated in breast cancer progression
J.Mol.Biol., 412, 2011
|
|
4WWQ
 
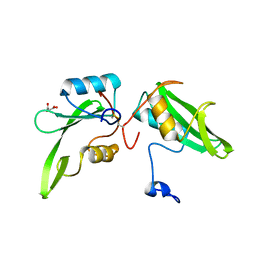 | | Apo structure of the Grb7 SH2 domain | | Descriptor: | Growth factor receptor-bound protein 7, MALONIC ACID | | Authors: | Watson, G.M, Ambaye, N.D, Wilce, M.C, Wilce, J.A. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic Peptides Incorporating Phosphotyrosine Mimetics as Potent and Specific Inhibitors of the Grb7 Breast Cancer Target.
J.Med.Chem., 58, 2015
|
|
4X6S
 
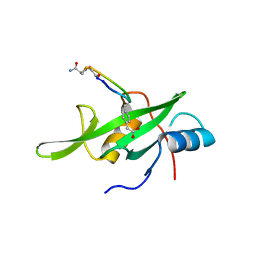 | | Grb7 SH2 domain with phosphotyrosine mimetic inhibitor peptide | | Descriptor: | Growth factor receptor-bound protein 7, Phosphotyrosine mimetic inhibitor peptide G7-TEM1 | | Authors: | Watson, G.M, Panjikar, S, Wilce, M.C, Wilce, J.A. | | Deposit date: | 2014-12-09 | | Release date: | 2015-09-23 | | Last modified: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Cyclic Peptides Incorporating Phosphotyrosine Mimetics as Potent and Specific Inhibitors of the Grb7 Breast Cancer Target.
J.Med.Chem., 58, 2015
|
|
5ITH
 
 | |
7MP3
 
 | |
1ZTG
 
 | | human alpha polyC binding protein KH1 | | Descriptor: | 5'-D(P*CP*CP*CP*TP*CP*CP*CP*T)-3', POLY(RC)-BINDING PROTEIN 1 | | Authors: | Sidiqi, M, Wilce, J.A, Barker, A, Schmidgerger, J, Leedman, P.J, Wilce, M.C.J. | | Deposit date: | 2005-05-27 | | Release date: | 2006-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Contribution of the first K-homology domain of poly(C)-binding protein 1 to its affinity and specificity for C-rich oligonucleotides
Nucleic Acids Res., 40, 2012
|
|
5D0J
 
 | | Grb7 SH2 with inhibitor peptide | | Descriptor: | G7-TEdFP peptide, Growth factor receptor-bound protein 7, PHOSPHATE ION | | Authors: | Gunzburg, M.J, Watson, G.M, Wilce, J.A, Wilce, M.C.J. | | Deposit date: | 2015-08-03 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
5EEL
 
 | | Grb7 SH2 with bicyclic peptide inhibitor | | Descriptor: | Bicyclic Peptide Inhibitor, FORMIC ACID, Growth factor receptor-bound protein 7, ... | | Authors: | Watson, G.M, Gunzburg, M.J, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2015-10-23 | | Release date: | 2016-06-15 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
4J1Y
 
 | | The X-ray crystal structure of human complement protease C1s zymogen | | Descriptor: | Complement C1s subcomponent | | Authors: | Perry, A.J, Wijeyewickrema, L.C, Wilmann, P.G, Gunzburg, M.J, D'Andrea, L, Irving, J.A, Pang, S.S, Duncan, R.C, Wilce, J.A, Whisstock, J.C, Pike, R.N. | | Deposit date: | 2013-02-03 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6645 Å) | | Cite: | A Molecular Switch Governs the Interaction between the Human Complement Protease C1s and Its Substrate, Complement C4.
J.Biol.Chem., 288, 2013
|
|
5U1Q
 
 | | Grb7-SH2 with bicyclic peptide inhibitor | | Descriptor: | CHLORIDE ION, Growth factor receptor-bound protein 7, LYS-PHE-GLU-GLY-TYR-ASP-ASN-GLU-CST | | Authors: | Watson, G.M, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
5TYI
 
 | | Grb7 SH2 with bicyclic peptide containing pY mimetic | | Descriptor: | Growth factor receptor-bound protein 7, Peptide inhibitor | | Authors: | Watson, G.M, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2016-11-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
8SUC
 
 | | NHL-2 NHL domain | | Descriptor: | MAGNESIUM ION, NHL (Ring finger b-box coiled coil) domain containing protein | | Authors: | Colson, R.N, Wilce, J.A. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | NHL-2 NHL domain
To Be Published
|
|
5U06
 
 | | Grb7-SH2 with bicyclic peptide inhibitor containing a pY mimetic | | Descriptor: | Growth factor receptor-bound protein 7, POTASSIUM ION, bicyclic peptide inhibitor: LYS-PHE-GLU-GLY-CMF-ASP-ASN-GLU-CST | | Authors: | Watson, G.M, Wilce, J.A. | | Deposit date: | 2016-11-22 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
2QMS
 
 | | Crystal structure of a signaling molecule | | Descriptor: | Growth factor receptor-bound protein 7, SULFATE ION | | Authors: | Porter, C.J, Wilce, M.C, Wilce, J.A. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Grb7 SH2 domain structure and interactions with a cyclic peptide inhibitor of cancer cell migration and proliferation.
Bmc Struct.Biol., 7, 2007
|
|
2EFW
 
 | | Crystal structure of the RTP:nRB complex from Bacillus subtilis | | Descriptor: | DNA (5'-D(*DCP*DT*DAP*DTP*DGP*DTP*DAP*DCP*DCP*DAP*DAP*DAP*DTP*DGP*DTP*DTP*DCP*DAP*DGP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DCP*DTP*DGP*DAP*DAP*DCP*DAP*DTP*DTP*DTP*DGP*DGP*DTP*DAP*DCP*DAP*DTP*DAP*DG)-3'), Replication termination protein | | Authors: | Vivian, J.P, Porter, C.J, Wilce, J.A, Wilce, M.C.J. | | Deposit date: | 2007-02-26 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An asymmetric structure of the Bacillus subtilis replication terminator protein in complex with DNA
J.Mol.Biol., 370, 2007
|
|
2DQR
 
 | |
1WVN
 
 | |
1J0R
 
 | | Crystal structure of the replication termination protein mutant C110S | | Descriptor: | replication termination protein | | Authors: | Vivian, J.P, Hastings, A.F, Duggin, I.G, Wake, R.G, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2002-11-20 | | Release date: | 2003-11-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The impact of single cysteine residue mutations on the replication terminator protein
Biochem.Biophys.Res.Commun., 310, 2003
|
|
3VKE
 
 | |
1WO3
 
 | | Solution structure of Minimal Mutant 1 (MM1): Multiple alanine mutant of non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO6
 
 | | Solution structure of Designed Functional Finger 5 (DFF5): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO4
 
 | | Solution structure of Minimal Mutant 2 (MM2): Multiple alanine mutant of non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO5
 
 | | Solution structure of Designed Functional Finger 2 (DFF2): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO7
 
 | | Solution structure of Designed Functional Finger 7 (DFF7): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
