4J0J
 
 | | Tannin acyl hydrolase in complex with ethyl 3,5-dihydroxybenzoate | | 分子名称: | Tannase, ethyl 3,5-dihydroxybenzoate | | 著者 | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | 登録日 | 2013-01-31 | | 公開日 | 2013-05-22 | | 最終更新日 | 2022-08-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4J0C
 
 | | tannin acyl hydrolase from Lactobacillus plantarum (native structure) | | 分子名称: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Tannase | | 著者 | Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | 登録日 | 2013-01-30 | | 公開日 | 2013-05-22 | | 最終更新日 | 2022-08-24 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4J0K
 
 | | Tannin acyl hydrolase in complex with ethyl gallate | | 分子名称: | DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | 著者 | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | 登録日 | 2013-01-31 | | 公開日 | 2013-05-22 | | 最終更新日 | 2022-08-24 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4J0D
 
 | | tannin acyl hydrolase from Lactobacillus plantarum (Cadmium) | | 分子名称: | CADMIUM ION, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | 著者 | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | 登録日 | 2013-01-30 | | 公開日 | 2013-05-22 | | 最終更新日 | 2022-08-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4J0H
 
 | | Tannin acyl hydrolase in complex with gallic acid | | 分子名称: | 3,4,5-trihydroxybenzoic acid, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | 著者 | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | 登録日 | 2013-01-30 | | 公開日 | 2013-05-22 | | 最終更新日 | 2022-08-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4JUI
 
 | | crystal structure of tannase from from Lactobacillus plantarum | | 分子名称: | DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | 著者 | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, McKinstry, W.J, Chen, Q. | | 登録日 | 2013-03-24 | | 公開日 | 2013-05-22 | | 最終更新日 | 2022-08-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
3ULX
 
 | |
6M71
 
 | | SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors | | 分子名称: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase | | 著者 | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | 登録日 | 2020-03-16 | | 公開日 | 2020-04-01 | | 最終更新日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
7BOK
 
 | | Cryo-EM structure of the encapsulated DyP-type peroxidase from Mycobacterium smegmatis | | 分子名称: | Dyp-type peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tang, Y.T, Mu, A, Gong, H.R, Wang, Q, Rao, Z.H. | | 登録日 | 2020-03-19 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BOJ
 
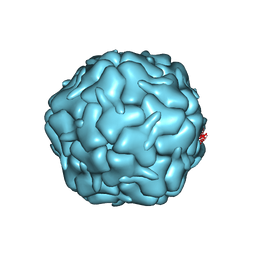 | | Cryo-EM structure of the encapsulin shell from Mycobacterium smegmatis | | 分子名称: | 29 kDa antigen Cfp29 | | 著者 | Tang, Y.T, Mu, A, Gong, H.R, Wang, Q, Rao, Z.H. | | 登録日 | 2020-03-19 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3DP4
 
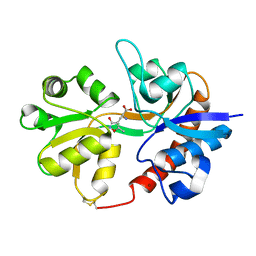 | | Crystal structure of the binding domain of the AMPA subunit GluR3 bound to AMPA | | 分子名称: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, Glutamate receptor 3, ZINC ION | | 著者 | Ahmed, A.H, Wang, Q, Sondermann, H, Oswald, R.E. | | 登録日 | 2008-07-07 | | 公開日 | 2008-11-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Structure of the S1S2 glutamate binding domain of GLuR3.
Proteins, 75, 2008
|
|
3DP6
 
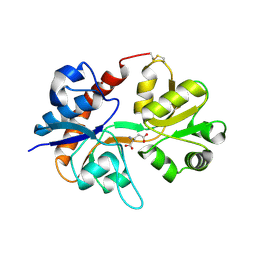 | | Crystal structure of the binding domain of the AMPA subunit GluR2 bound to glutamate | | 分子名称: | GLUTAMIC ACID, Glutamate receptor 2, ZINC ION | | 著者 | Ahmed, A.H, Wang, Q, Sondermann, H, Oswald, R.E. | | 登録日 | 2008-07-07 | | 公開日 | 2008-11-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure of the S1S2 glutamate binding domain of GLuR3.
Proteins, 75, 2008
|
|
3DLN
 
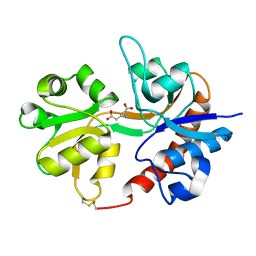 | | Crystal structure of the binding domain of the AMPA subunit GluR3 bound to glutamate | | 分子名称: | GLUTAMIC ACID, Glutamate receptor 3, ZINC ION | | 著者 | Ahmed, A.H, Wang, Q, Sondermann, H, Oswald, R.E. | | 登録日 | 2008-06-27 | | 公開日 | 2008-11-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Structure of the S1S2 glutamate binding domain of GLuR3.
Proteins, 75, 2008
|
|
7U4M
 
 | | Crystal structure of human GPX4-U46C in complex with LOC1886 | | 分子名称: | 1,2-ETHANEDIOL, 4-methoxy-1H-indole-2-carbaldehyde, Phospholipid hydroperoxide glutathione peroxidase | | 著者 | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | 登録日 | 2022-02-28 | | 公開日 | 2022-12-07 | | 最終更新日 | 2022-12-28 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4K
 
 | | Crystal structure of human GPX4-U46C-R152H in complex with ML162 | | 分子名称: | 1,2-ETHANEDIOL, 2-chloro-N-(3-chloro-4-methoxyphenyl)-N-[(1R)-2-oxo-2-[(2-phenylethyl)amino]-1-(thiophen-2-yl)ethyl]acetamide, Phospholipid hydroperoxide glutathione peroxidase | | 著者 | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | 登録日 | 2022-02-28 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4J
 
 | | Crystal structure of human GPX4-U46C-R152H in complex with TMT10 | | 分子名称: | Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION, ~{N}-(3-chloranyl-4-methoxy-phenyl)ethanamide | | 著者 | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | 登録日 | 2022-02-28 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4I
 
 | | Crystal structure of human GPX4-U46C-R152H in complex with CDS9 | | 分子名称: | 2-bromo-N-[(thiophen-2-yl)methyl]acetamide, Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION | | 著者 | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | 登録日 | 2022-02-28 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4N
 
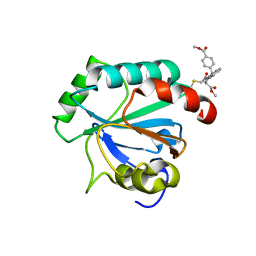 | | Crystal structure of human GPX4-U46C in complex with RSL3 | | 分子名称: | Phospholipid hydroperoxide glutathione peroxidase, methyl (1S,3R)-2-(chloroacetyl)-1-[4-(methoxycarbonyl)phenyl]-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxylate | | 著者 | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | 登録日 | 2022-02-28 | | 公開日 | 2022-12-07 | | 最終更新日 | 2022-12-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4L
 
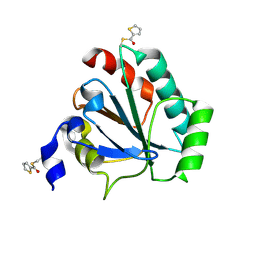 | | Crystal structure of human GPX4-U46C in complex with MAC-5576 | | 分子名称: | Phospholipid hydroperoxide glutathione peroxidase, thiophene-2-carbaldehyde | | 著者 | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | 登録日 | 2022-02-28 | | 公開日 | 2022-12-07 | | 最終更新日 | 2022-12-28 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
3FUS
 
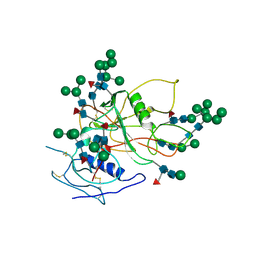 | | Improved Structure of the Unliganded Simian Immunodeficiency Virus gp120 Core | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Chen, X, Poon, B, Wang, Q, Ma, J. | | 登録日 | 2009-01-14 | | 公開日 | 2009-06-30 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | Structural improvement of unliganded simian immunodeficiency virus gp120 core by normal-mode-based X-ray crystallographic refinement.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3LUT
 
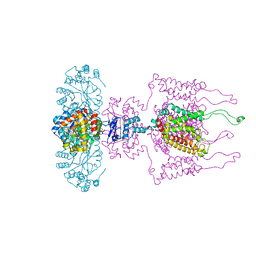 | | A Structural Model for the Full-length Shaker Potassium Channel Kv1.2 | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 2, ... | | 著者 | Chen, X, Ni, F, Wang, Q, Ma, J. | | 登録日 | 2010-02-18 | | 公開日 | 2010-06-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4DUI
 
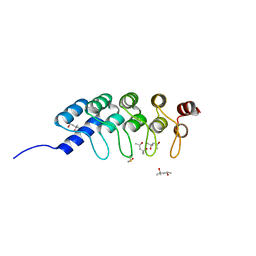 | | DARPIN D1 binding to tubulin beta chain (not in complex) | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, DESIGNED ANKYRIN REPEAT PROTEIN (DARPIN) D1 | | 著者 | Pecqueur, L, Duellberg, C, Dreier, B, Wang, Q, Jiang, C, Pluckthun, A, Surrey, T, Gigant, B, Knossow, M. | | 登録日 | 2012-02-22 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | An Anti-Tubulin Darpin Caps the Microtubule Plus-End
To be Published
|
|
4E41
 
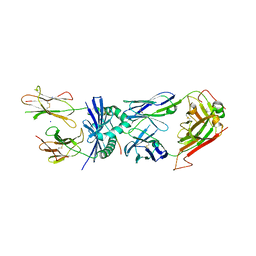 | | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4 | | 分子名称: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-1 beta chain, ... | | 著者 | Deng, L, Langley, R.J, Wang, Q, Topalian, S.L, Mariuzza, R.A. | | 登録日 | 2012-03-11 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4
Proc.Natl.Acad.Sci.USA, 2012
|
|
4E42
 
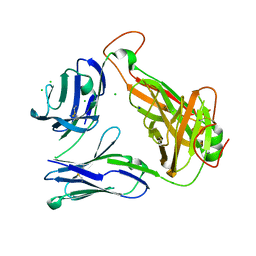 | | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4 | | 分子名称: | CHLORIDE ION, NITRATE ION, SODIUM ION, ... | | 著者 | Deng, L, Langley, R.J, Wang, Q, Topalian, S.L, Mariuzza, R.A. | | 登録日 | 2012-03-11 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4
Proc.Natl.Acad.Sci.USA, 2012
|
|
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | 分子名称: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | 著者 | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
