6EN3
 
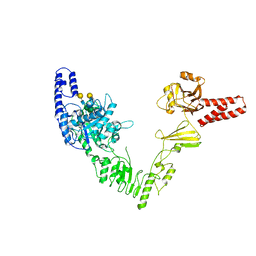 | | Crystal structure of full length EndoS from Streptococcus pyogenes in complex with G2 oligosaccharide. | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F2,Multifunctional-autoprocessing repeats-in-toxin, NICKEL (II) ION, ... | | Authors: | Trastoy, B, Klontz, E.H, Orwenyo, J, Marina, A, Wang, L.X, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2017-10-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structural basis for the recognition of complex-type N-glycans by Endoglycosidase S.
Nat Commun, 9, 2018
|
|
3FHA
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, GLYCEROL, ... | | Authors: | Yin, J, Li, L, Shaw, N, Li, Y, Song, J.K, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.C, Wang, L.X, Wang, P, Liu, Z.J. | | Deposit date: | 2008-12-09 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A.
Plos One, 4, 2009
|
|
4Q6Y
 
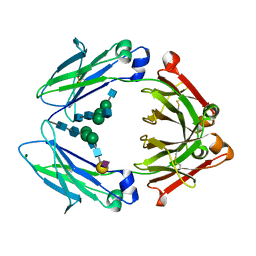 | | Crystal structure of a chemoenzymatic glycoengineered disialylated Fc (di-sFc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ahmed, A.A, Giddens, J, Pincetic, A, Lomino, J.V, Ravetch, J.V, Wang, L.X, Bjorkman, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural characterization of anti-inflammatory immunoglobulin g fc proteins.
J.Mol.Biol., 426, 2014
|
|
4Q74
 
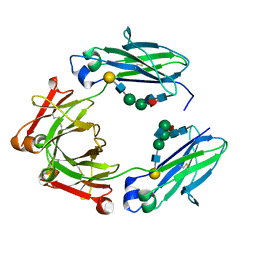 | | F241A Fc | | Descriptor: | Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ahmed, A.A, Giddens, J, Pincetic, A, Lomino, J.V, Ravetch, J.V, Wang, L.X, Bjorkman, P.J. | | Deposit date: | 2014-04-24 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural characterization of anti-inflammatory immunoglobulin g fc proteins.
J.Mol.Biol., 426, 2014
|
|
4Q7D
 
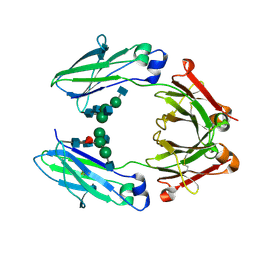 | | Wild type Fc (wtFc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Ahmed, A.A, Giddens, J, Pincetic, A, Lomino, J.V, Ravetch, J.V, Wang, L.X, Bjorkman, P.J. | | Deposit date: | 2014-04-24 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of anti-inflammatory immunoglobulin g fc proteins.
J.Mol.Biol., 426, 2014
|
|
6MDS
 
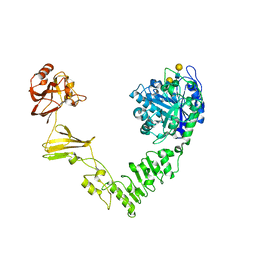 | | Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) with complex biantennary glycan | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Klontz, E.H, Trastoy, B, Orwenyo, J, Wang, L.X, Guerin, M.E, Sundberg, E.J. | | Deposit date: | 2018-09-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis of Broad SpectrumN-Glycan Specificity and Processing of Therapeutic IgG Monoclonal Antibodies by Endoglycosidase S2.
ACS Cent Sci, 5, 2019
|
|
6MDV
 
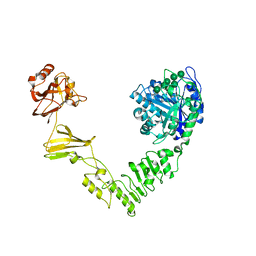 | | Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) with high-mannose glycan | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Klontz, E.H, Trastoy, B, Orwenyo, J, Wang, L.X, Guerin, M.E, Sundberg, E.J. | | Deposit date: | 2018-09-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis of Broad SpectrumN-Glycan Specificity and Processing of Therapeutic IgG Monoclonal Antibodies by Endoglycosidase S2.
ACS Cent Sci, 5, 2019
|
|
6T8K
 
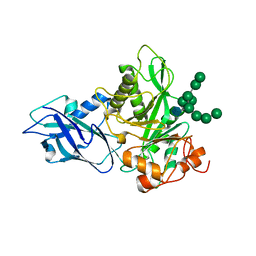 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 in complex with Man9GlcNAc product in P1 | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F1, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
6TCV
 
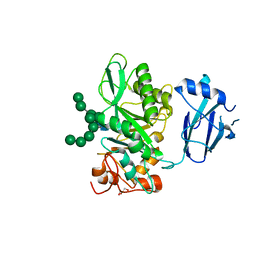 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 in complex with Man9GlcNAc2Asn substrate | | Descriptor: | ASPARAGINE, Endo-beta-N-acetylglucosaminidase F1, GLYCEROL, ... | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-11-06 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.311 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
6T8I
 
 | | Crystal structure of wild type EndoBT-3987 from Bacteroides thetaiotamicron VPI-5482 | | Descriptor: | Endo-beta-N-acetylglucosaminidase F1, GLYCEROL | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
6T8L
 
 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 with Man9GlcNAc product in P212121 | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F1, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
6TCW
 
 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 with Man5GlcNAc product | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F1, GLYCEROL, ... | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-11-06 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
7NWF
 
 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 in complex with hybrid-type glycan (GalGlcNAcMan5GlcNAc) product | | Descriptor: | Endo-beta-N-acetylglucosaminidase F1, GLYCEROL, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Trastoy, B, Du, J.J, Garcia-Alija, M, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2021-03-16 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GH18 endo-beta-N-acetylglucosaminidases use distinct mechanisms to process hybrid-type N-linked glycans.
J.Biol.Chem., 297, 2021
|
|
8A64
 
 | | cryoEM structure of the catalytically inactive EndoS from S. pyogenes in complex with the Fc region of immunoglobulin G1. | | Descriptor: | Endo-beta-N-acetylglucosaminidase F2, Immunoglobulin gamma-1 heavy chain, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Trastoy, B, Cifuente, J.O, Du, J.J, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2022-06-16 | | Release date: | 2023-04-05 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism of antibody-specific deglycosylation and immune evasion by Streptococcal IgG-specific endoglycosidases.
Nat Commun, 14, 2023
|
|
6X5S
 
 | | Human Alpha-1,6-fucosyltransferase (FUT8) bound to GDP and A3'-Asn | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A3'-Asn, Alpha-(1,6)-fucosyltransferase, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
J.Biol.Chem., 295, 2020
|
|
6X5R
 
 | | Human Alpha-1,6-fucosyltransferase (FUT8) bound to GDP and A2-Asn | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A2-Asn, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
J.Biol.Chem., 295, 2020
|
|
6X5T
 
 | | Human Alpha-1,6-fucosyltransferase (FUT8) bound to GDP and A3-Asn | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-(1,6)-fucosyltransferase, GLYCEROL, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
J.Biol.Chem., 295, 2020
|
|
8GQE
 
 | | Crystal structure of the W285A mutant of UVR8 in complex with RUP2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ultraviolet-B receptor UVR8, WD repeat-containing protein RUP2 | | Authors: | Wang, Y.D, Wang, L.X, Guan, Z.Y, chang, H.F, Yin, P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RUP2 facilitates UVR8 redimerization via two interfaces.
Plant Commun., 4, 2023
|
|
6OHE
 
 | |
6E20
 
 | | Crystal structure of the Dario rerio galectin-1-L2 | | Descriptor: | Galectin, MAGNESIUM ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Ghosh, A, Bianchet, M.A. | | Deposit date: | 2018-07-10 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the zebrafish galectin-1-L2 and model of its interaction with the infectious hematopoietic necrosis virus (IHNV) envelope glycoprotein.
Glycobiology, 29, 2019
|
|
3FHQ
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Endo-beta-N-acetylglucosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose | | Authors: | Jie, Y, Li, L, Shaw, N, Li, Y, Song, J, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.-C, Wang, L.-X, Wang, P, Liu, Z.-J. | | Deposit date: | 2008-12-10 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A
Plos One, 4, 2009
|
|
4DQO
 
 | | Crystal Structure of PG16 Fab in Complex with V1V2 Region from HIV-1 strain ZM109 | | Descriptor: | 1FD6-V1V2 scaffold ZM109 HIV-1 strain, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG16 Fab Heavy Chain, ... | | Authors: | Pancera, M, McLellan, J.S, Kwong, P.D. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6X5H
 
 | | Human Alpha-1,6-fucosyltransferase (FUT8) bound to GDP | | Descriptor: | 1,2-ETHANEDIOL, Alpha-(1,6)-fucosyltransferase, GLYCEROL, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
J.Biol.Chem., 295, 2020
|
|
6X5U
 
 | | Human Alpha-1,6-fucosyltransferase (FUT8) bound to GDP and NM5M2-Asn | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
J.Biol.Chem., 295, 2020
|
|
3IF1
 
 | | Crystal structure of 237mAb in complex with a GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Immunoglobulin heavy chain (IgG2a), Immunoglobulin light chain (IgG2a), ... | | Authors: | Brooks, C.L, Evans, S.V, Borisova, S.N. | | Deposit date: | 2009-07-23 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Antibody recognition of a unique tumor-specific glycopeptide antigen.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
