3PP7
 
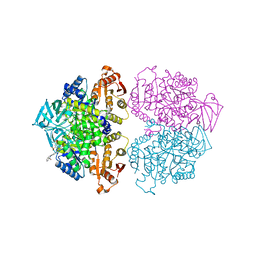 | | Crystal structure of Leishmania mexicana pyruvate kinase in complex with the drug suramin, an inhibitor of glycolysis. | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Morgan, H.P, Auld, D.S, McNae, I.W, Nowicki, M.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2010-11-24 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The trypanocidal drug suramin and other trypan blue mimetics are inhibitors of pyruvate kinases and bind to the adenosine site.
J.Biol.Chem., 286, 2011
|
|
3QV9
 
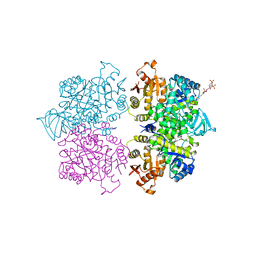 | | Crystal structure of Trypanosoma cruzi pyruvate kinase(TcPYK)in complex with ponceau S. | | Descriptor: | 3-hydroxy-4-[(E)-{2-sulfo-4-[(E)-(4-sulfophenyl)diazenyl]phenyl}diazenyl]naphthalene-2,7-disulfonic acid, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Morgan, H.P, Auld, D.S, McNae, I.W, Nowicki, M.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-02-25 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The trypanocidal drug suramin and other trypan blue mimetics are inhibitors of pyruvate kinases and bind to the adenosine site.
J.Biol.Chem., 286, 2011
|
|
3QV7
 
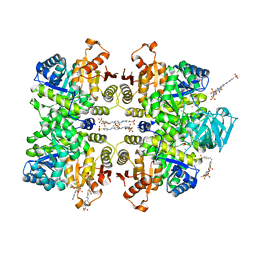 | | Crystal structure of Leishmania mexicana pyruvate kinase(LmPYK)in complex with ponceau S and acid blue 25. | | Descriptor: | 3-hydroxy-4-[(E)-{2-sulfo-4-[(E)-(4-sulfophenyl)diazenyl]phenyl}diazenyl]naphthalene-2,7-disulfonic acid, 9,10-dioxo-4-(phenylamino)-9,10-dihydroanthracene-2-sulfonic acid, POTASSIUM ION, ... | | Authors: | Morgan, H.P, Auld, D.S, McNae, I.W, Nowicki, M.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-02-25 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The trypanocidal drug suramin and other trypan blue mimetics are inhibitors of pyruvate kinases and bind to the adenosine site.
J.Biol.Chem., 286, 2011
|
|
3QV6
 
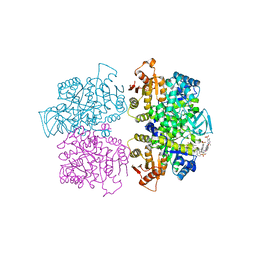 | | Crystal structure of Leishmania mexicana pyruvate kinase(LmPYK)in complex with acid blue 80. | | Descriptor: | 3-({4-[(2,4-dimethyl-5-sulfophenyl)amino]-9,10-dioxo-9,10-dihydroanthracen-1-yl}amino)-2,4,6-trimethylbenzenesulfonic acid, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Morgan, H.P, Auld, D.S, McNae, I.W, Nowicki, M.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-02-25 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The trypanocidal drug suramin and other trypan blue mimetics are inhibitors of pyruvate kinases and bind to the adenosine site.
J.Biol.Chem., 286, 2011
|
|
3QV8
 
 | | Crystal structure of Leishmania mexicana pyruvate kinase(LmPYK)in complex with benzothiazole-2,5-disulfonic acid. | | Descriptor: | 1,3-benzothiazole-2,5-disulfonic acid, PHOSPHATE ION, Pyruvate kinase | | Authors: | Morgan, H.P, Auld, D.S, McNae, I.W, Nowicki, M.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-02-25 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The trypanocidal drug suramin and other trypan blue mimetics are inhibitors of pyruvate kinases and bind to the adenosine site.
J.Biol.Chem., 286, 2011
|
|
2V8W
 
 | | Crystallographic and mass spectrometric characterisation of eIF4E with N7-cap derivatives | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E-BINDING PROTEIN 1, [[(2R,3S,4R,5R)-5-(6-AMINO-3-METHYL-4-OXO-5H-IMIDAZO[4,5-C]PYRIDIN-1-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHOXY-HYDROXY-PHOSPHORYL] PHOSPHONO HYDROGEN PHOSPHATE | | Authors: | Brown, C.J, Mcnae, I, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2007-08-16 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic and Mass Spectrometric Characterisation of Eif4E with N(7)-Alkylated CAP Derivatives.
J.Mol.Biol., 372, 2007
|
|
2V8Y
 
 | | Crystallographic and mass spectrometric characterisation of eIF4E with N7-cap derivatives | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E-BINDING PROTEIN 1, P-FLUORO-7-BENZYL GUANINE MONOPHOSPHATE | | Authors: | Brown, C.J, Mcnae, I, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2007-08-16 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic and Mass Spectrometric Characterisation of Eif4E with N(7)-Alkylated CAP Derivatives.
J.Mol.Biol., 372, 2007
|
|
2V8X
 
 | | Crystallographic and mass spectrometric characterisation of eIF4E with N7-cap derivatives | | Descriptor: | 7-BENZYL GUANINE MONOPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E-BINDING PROTEIN 1 | | Authors: | Brown, C.J, Mcnae, I, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2007-08-16 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic and Mass Spectrometric Characterisation of Eif4E with N(7)-Alkylated CAP Derivatives.
J.Mol.Biol., 372, 2007
|
|
2W82
 
 | | The structure of ArdA | | Descriptor: | ORF18 | | Authors: | McMahon, S.A, Roberts, G.A, Carter, L.G, Cooper, L.P, Liu, H, White, J.H, Johnson, K.A, Sanghvi, B, Oke, M, Walkinshaw, M.D, Blakely, G, Naismith, J.H, Dryden, D.T.F. | | Deposit date: | 2009-01-08 | | Release date: | 2009-01-27 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Extensive DNA Mimicry by the Arda Anti-Restriction Protein and its Role in the Spread of Antibiotic Resistance.
Nucleic Acids Res., 37, 2009
|
|
2W97
 
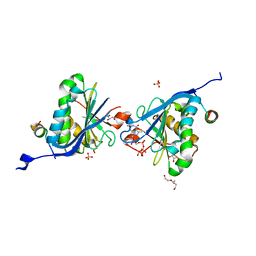 | | Crystal Structure of eIF4E Bound to Glycerol and eIF4G1 peptide | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4 GAMMA 1, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, GLYCEROL, ... | | Authors: | Brown, C.J, Verma, C.S, Walkinshaw, M.D, Lane, D.P. | | Deposit date: | 2009-01-22 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystallization of eIF4E complexed with eIF4GI peptide and glycerol reveals distinct structural differences around the cap-binding site.
Cell Cycle, 8, 2009
|
|
1E8K
 
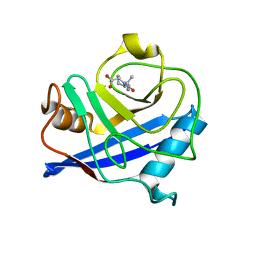 | | Cyclophilin 3 Complexed With Dipeptide Ala-Pro | | Descriptor: | ALANINE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE 3, PROLINE | | Authors: | Wu, S.Y, Dornan, J, Kontopidis, G, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 2000-09-25 | | Release date: | 2001-09-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The First Direct Determination of a Ligand Binding Constant in Protein Crystals
Angew.Chem.Int.Ed.Engl., 40, 2001
|
|
4KS0
 
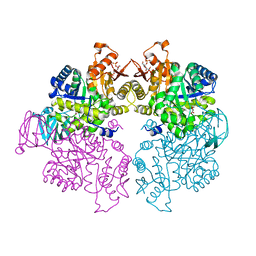 | | Pyruvate kinase (PYK) from Trypanosoma cruzi in the presence of Magnesium, oxalate and F26BP | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Morgan, H.P, Zhong, W, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2013-05-17 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of pyruvate kinases display evolutionarily divergent allosteric strategies.
R Soc Open Sci, 1, 2014
|
|
4N1Q
 
 | |
4N1N
 
 | |
4N1P
 
 | |
4N1S
 
 | |
4N1O
 
 | |
4N1R
 
 | |
4N1M
 
 | |
1E39
 
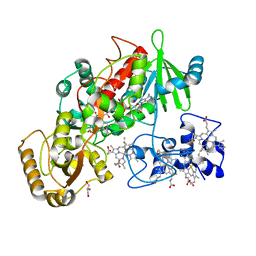 | | Flavocytochrome C3 from Shewanella frigidimarina histidine 365 mutated to alanine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FUMARATE REDUCTASE FLAVOPROTEIN SUBUNIT, FUMARIC ACID, ... | | Authors: | Doherty, M.K, Pealing, S.L, Miles, C.S, Moysey, R, Taylor, P, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2000-06-07 | | Release date: | 2000-09-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of the Active Site Acid/Base Catalyst in a Bacterial Fumarate Reductase: A Kinetic and Crystallographic Study
Biochemistry, 39, 2000
|
|
1YQO
 
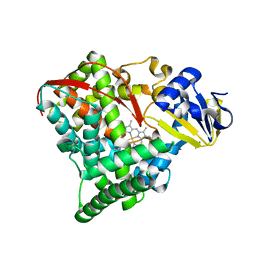 | | T268A mutant heme domain of flavocytochrome P450 BM3 | | Descriptor: | Bifunctional P-450:NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Clark, J.P, Miles, C.S, Mowat, C.G, Walkinshaw, M.D, Reid, G.A, Daff, S.N, Chapman, S.K. | | Deposit date: | 2005-02-02 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of Thr268 and Phe393 in cytochrome P450 BM3.
J.Inorg.Biochem., 100, 2006
|
|
2A0C
 
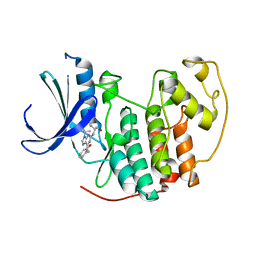 | | Human CDK2 in complex with olomoucine II, a novel 2,6,9-trisubstituted purine cyclin-dependent kinase inhibitor | | Descriptor: | 2-{[(2-{[(1R)-1-(HYDROXYMETHYL)PROPYL]AMINO}-9-ISOPROPYL-9H-PURIN-6-YL)AMINO]METHYL}PHENOL, Cell division protein kinase 2 | | Authors: | Krystof, V, McNae, I.W, Walkinshaw, M.D, Fischer, P.M, Muller, P, Vojtesek, B, Orsag, M, Havlicek, L, Strnad, M. | | Deposit date: | 2005-06-16 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antiproliferative activity of olomoucine II, a novel 2,6,9-trisubstituted purine cyclin-dependent kinase inhibitor
Cell.Mol.Life Sci., 62, 2005
|
|
2B7R
 
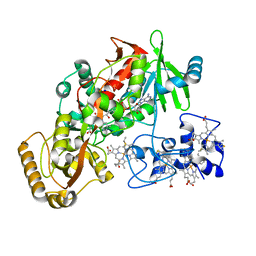 | | Structure of E378D mutant flavocytochrome c3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, Fumarate reductase flavoprotein subunit, ... | | Authors: | Pankhurst, K.L, Mowat, C.G, Rothery, E.L, Miles, C.S, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2005-10-05 | | Release date: | 2006-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Proton Delivery Pathway in the Soluble Fumarate Reductase from Shewanella frigidimarina.
J.Biol.Chem., 281, 2006
|
|
2B7S
 
 | | R381K mutant of flavocytochrome c3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, Fumarate reductase flavoprotein subunit, ... | | Authors: | Pankhurst, K.L, Mowat, C.G, Rothery, E.L, Miles, C.S, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2005-10-05 | | Release date: | 2006-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A Proton Delivery Pathway in the Soluble Fumarate Reductase from Shewanella frigidimarina.
J.Biol.Chem., 281, 2006
|
|
2BLC
 
 | | SP21 double mutant P. vivax Dihydrofolate reductase in complex with des-chloropyrimethamine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-ETHYL-5-PHENYLPYRIMIDINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE-THYMIDYLATE SYNTHASE, ... | | Authors: | Kongsaeree, P, Khongsuk, P, Leartsakulpanich, U, Chitnumsub, P, Tarnchompoo, B, Walkinshaw, M.D, Yuthavong, Y. | | Deposit date: | 2005-03-03 | | Release date: | 2005-09-07 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Dihydrofolate Reductase from Plasmodium Vivax: Pyrimethamine Displacement Linked with Mutation-Induced Resistance.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
