1GQE
 
 | |
2ANJ
 
 | | Crystal Structure of the Glur2 Ligand Binding Core (S1S2J-Y450W) Mutant in Complex With the Partial Agonist Kainic Acid at 2.1 A Resolution | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | 著者 | Holm, M.M, Naur, P, Vestergaard, B, Geballe, M.T, Gajhede, M, Kastrup, J.S, Traynelis, S.F, Egebjerg, J. | | 登録日 | 2005-08-11 | | 公開日 | 2005-08-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A Binding Site Tyrosine Shapes Desensitization Kinetics and Agonist Potency at GluR2: a mutagenic, kinetic, and crystallographic study
J.Biol.Chem., 280, 2005
|
|
1MVR
 
 | | Decoding Center & Peptidyl transferase center from the X-ray structure of the Thermus thermophilus 70S ribosome, aligned to the low resolution Cryo-EM map of E.coli 70S Ribosome | | 分子名称: | 30S RIBOSOMAL PROTEIN S12, 50S ribosomal protein L11, Helix 34 of 16S rRNA, ... | | 著者 | Rawat, U.B, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | 登録日 | 2002-09-26 | | 公開日 | 2003-04-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (12.8 Å) | | 主引用文献 | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
1ML5
 
 | | Structure of the E. coli ribosomal termination complex with release factor 2 | | 分子名称: | 30S 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | 著者 | Klaholz, B.P, Pape, T, Zavialov, A.V, Myasnikov, A.G, Orlova, E.V, Vestergaard, B, Ehrenberg, M, van Heel, M. | | 登録日 | 2002-08-30 | | 公開日 | 2003-01-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (14 Å) | | 主引用文献 | Structure of the Escherichia coli ribosomal termination complex with release factor 2
Nature, 421, 2003
|
|
1MI6
 
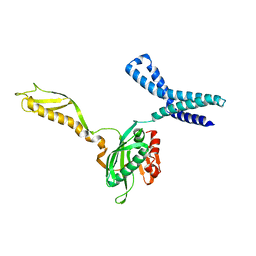 | | Docking of the modified RF2 X-ray structure into the Low Resolution Cryo-EM map of RF2 E.coli 70S Ribosome | | 分子名称: | peptide chain release factor RF-2 | | 著者 | Rawat, U.B.S, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | 登録日 | 2002-08-22 | | 公開日 | 2003-01-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (12.8 Å) | | 主引用文献 | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
2V3U
 
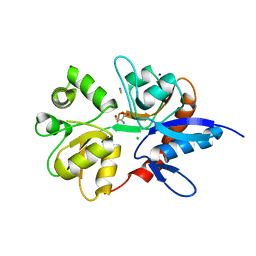 | | Structure of the ligand-binding core of the ionotropic glutamate receptor-like GluRdelta2 in complex with D-serine | | 分子名称: | CHLORIDE ION, D-SERINE, GLUTAMATE RECEPTOR DELTA-2 SUBUNIT, ... | | 著者 | Naur, P, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | 登録日 | 2007-06-22 | | 公開日 | 2007-08-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Ionotropic Glutamate-Like Receptor {Delta}2 Binds D-Serine and Glycine.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
3BFT
 
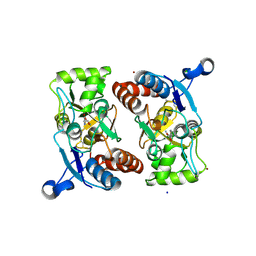 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (S)-TDPA at 2.25 A resolution | | 分子名称: | (2S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, CACODYLATE ION, CHLORIDE ION, ... | | 著者 | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | 登録日 | 2007-11-23 | | 公開日 | 2008-10-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
3BFU
 
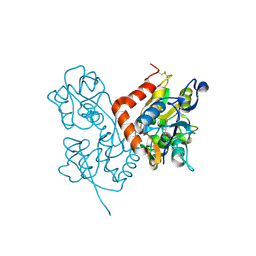 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (R)-TDPA at 1.95 A resolution | | 分子名称: | (2R)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, Glutamate receptor 2 | | 著者 | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | 登録日 | 2007-11-23 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
1SYI
 
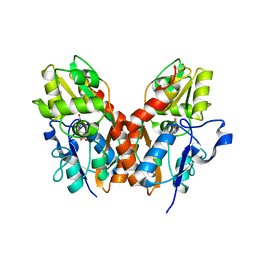 | | X-RAY STRUCTURE OF THE Y702F MUTANT OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 2.1 A RESOLUTION. | | 分子名称: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | 著者 | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | 登録日 | 2004-04-01 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
1SYH
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 1.85 A RESOLUTION. | | 分子名称: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | 著者 | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | 登録日 | 2004-04-01 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
1XHY
 
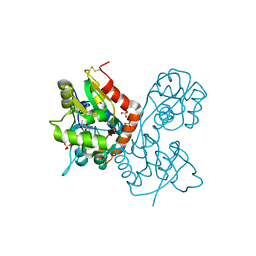 | | X-ray structure of the Y702F mutant of the GluR2 ligand-binding core (S1S2J) in complex with kainate at 1.85 A resolution | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor, SULFATE ION | | 著者 | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | 登録日 | 2004-09-21 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
1YCJ
 
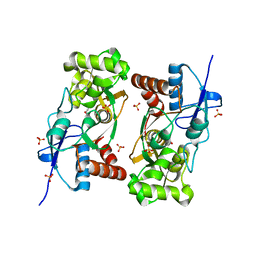 | | Crystal structure of the kainate receptor GluR5 ligand-binding core in complex with (S)-glutamate | | 分子名称: | GLUTAMIC ACID, Ionotropic glutamate receptor 5, SULFATE ION | | 著者 | Naur, P, Vestergaard, B, Skov, L.K, Egebjerg, J, Gajhede, M, Kastrup, J.S. | | 登録日 | 2004-12-22 | | 公開日 | 2005-02-01 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of the kainate receptor GluR5 ligand-binding core in complex with (S)-glutamate
Febs Lett., 579, 2005
|
|
2V3T
 
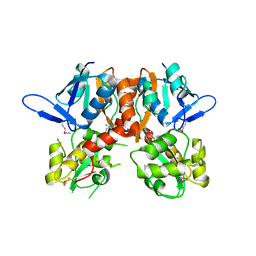 | | Structure of the ligand-binding core of the ionotropic glutamate receptor-like GluRdelta2 in the apo form | | 分子名称: | CALCIUM ION, GLUTAMATE RECEPTOR DELTA-2 SUBUNIT SYNONYM GLURDELTA2, GLUR DELTA-2 | | 著者 | Naur, P, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | 登録日 | 2007-06-22 | | 公開日 | 2007-08-07 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Ionotropic Glutamate-Like Receptor {Delta}2 Binds D-Serine and Glycine.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2JLL
 
 | | Crystal structure of NCAM2 IgIV-FN3II | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | 著者 | Kulahin, N, Rasmussen, K, Kristensen, O, Kastrup, J, Berezin, V, Bock, E, Walmod, P, Gajhede, M. | | 登録日 | 2008-09-10 | | 公開日 | 2009-11-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
4YMB
 
 | | Structure of the ligand-binding domain of GluK1 in complex with the antagonist CNG10111 | | 分子名称: | (3R,4S)-3-(3-carboxyphenyl)-4-propyl-L-proline, 1,2-ETHANEDIOL, ACETATE ION, ... | | 著者 | Moller, C, Tapken, D, Kastrup, J.S, Frydenvang, K. | | 登録日 | 2015-03-06 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structure-Activity Relationship Study of Ionotropic Glutamate Receptor Antagonist (2S,3R)-3-(3-Carboxyphenyl)pyrrolidine-2-carboxylic Acid.
J.Med.Chem., 58, 2015
|
|
3ZRT
 
 | |
2V5T
 
 | | Crystal structure of NCAM2 Ig2-3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NEURAL CELL ADHESION MOLECULE 2, ... | | 著者 | Kulahin, N, Rasmussen, K.K, Kristensen, O, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | 登録日 | 2007-07-10 | | 公開日 | 2008-07-29 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
2WIM
 
 | | Crystal structure of NCAM2 IG1-3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAL CELL ADHESION MOLECULE 2 | | 著者 | Kulahin, N, Kristensen, O, Rasmussen, K, Kastrup, J, Berezin, V, Bock, E, Walmod, P, Gajhede, M. | | 登録日 | 2009-05-13 | | 公開日 | 2010-08-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural model and trans-interaction of the entire ectodomain of the olfactory cell adhesion molecule.
Structure, 19, 2011
|
|
2XYC
 
 | | CRYSTAL STRUCTURE OF NCAM2 IGIV-FN3I | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NEURAL CELL ADHESION MOLECULE 2, ... | | 著者 | Kulahin, N, Rasmussen, K.K, Kristensen, O, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | 登録日 | 2010-11-17 | | 公開日 | 2011-02-23 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
2XY1
 
 | | CRYSTAL STRUCTURE OF NCAM2 IG3-4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NEURAL CELL ADHESION MOLECULE 2 | | 著者 | Kulahin, N, Rasmussen, K.K, Kristensen, O, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | 登録日 | 2010-11-12 | | 公開日 | 2011-02-23 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.979 Å) | | 主引用文献 | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
2XY2
 
 | | CRYSTAL STRUCTURE OF NCAM2 IG1-2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NEURAL CELL ADHESION MOLECULE 2 | | 著者 | Kulahin, N, Rasmussen, K.K, Kristensen, O, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | 登録日 | 2010-11-12 | | 公開日 | 2011-02-23 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
5M2V
 
 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with (2S,4R)-4-(2-carboxyphenoxy)pyrrolidine-2-carboxylic acid at 3.18 A resolution | | 分子名称: | (2~{S},4~{R})-4-(2-carboxyphenoxy)pyrrolidine-2-carboxylic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | 著者 | Frydenvang, K, Kastrup, J.S, Kristensen, C.M. | | 登録日 | 2016-10-13 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | Design and Synthesis of a Series of l-trans-4-Substituted Prolines as Selective Antagonists for the Ionotropic Glutamate Receptors Including Functional and X-ray Crystallographic Studies of New Subtype Selective Kainic Acid Receptor Subtype 1 (GluK1) Antagonist (2S,4R)-4-(2-Carboxyphenoxy)pyrrolidine-2-carboxylic Acid.
J. Med. Chem., 60, 2017
|
|
5MFW
 
 | | Crystal structure of the GluK1 ligand-binding domain in complex with kainate and BPAM-121 at 2.10 A resolution | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 7-chloro-4-(2-fluoroethyl)-2,3-dihydro-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, ... | | 著者 | Larsen, A.P, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2016-11-18 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification and Structure-Function Study of Positive Allosteric Modulators of Kainate Receptors.
Mol. Pharmacol., 91, 2017
|
|
5MFQ
 
 | | Crystal structure of the GluK1 ligand-binding domain in complex with kainate and BPAM-344 at 1.90 A resolution | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, ... | | 著者 | Larsen, A.P, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2016-11-18 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Identification and Structure-Function Study of Positive Allosteric Modulators of Kainate Receptors.
Mol. Pharmacol., 91, 2017
|
|
5MFV
 
 | | Crystal structure of the GluK1 ligand-binding domain in complex with kainate and BPAM-521 at 2.18 A resolution | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 4-Cyclopropyl-3,4-dihydro-7-hydroxy-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, ... | | 著者 | Larsen, A.P, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2016-11-18 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.181 Å) | | 主引用文献 | Identification and Structure-Function Study of Positive Allosteric Modulators of Kainate Receptors.
Mol. Pharmacol., 91, 2017
|
|
