1MSN
 
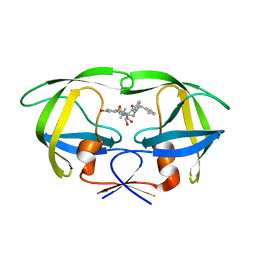 | | The HIV protease (mutant Q7K L33I L63I V82F I84V) complexed with KNI-764 (an inhibitor) | | Descriptor: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-19 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
1MRX
 
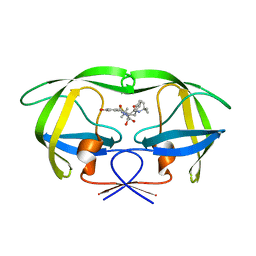 | | Structure of HIV protease (Mutant Q7K L33I L63I V82F I84V ) complexed with KNI-577 | | Descriptor: | (4R)-N-tert-butyl-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-1,3-thiazoli dine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-18 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
1MSM
 
 | | The HIV protease (mutant Q7K L33I L63I) complexed with KNI-764 (an inhibitor) | | Descriptor: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-19 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
1MRW
 
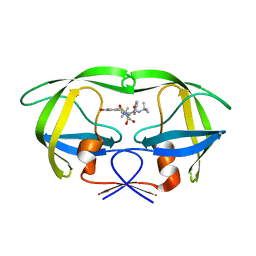 | | Structure of HIV protease (Mutant Q7K L33I L63I) complexed with KNI-577 | | Descriptor: | (4R)-N-tert-butyl-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-1,3-thiazoli dine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-18 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
4K09
 
 | |
4K06
 
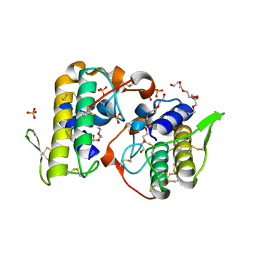 | | Crystal structure of MTX-II from Bothrops brazili venom complexed with polyethylene glycol | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, MTX-II, ... | | Authors: | Fernandes, C.A.H, Comparetti, E.J, Borges, R.J, Fontes, M.R.M. | | Deposit date: | 2013-04-03 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural bases for a complete myotoxic mechanism: Crystal structures of two non-catalytic phospholipases A2-like from Bothrops brazili venom.
Biochim.Biophys.Acta, 1834, 2013
|
|
5LJP
 
 | |
8XF7
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with ferrioxamine E bound | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, 1,2-ETHANEDIOL, FE (III) ION, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XEU
 
 | |
8XFA
 
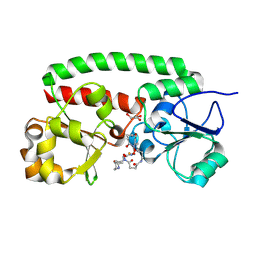 | | Structure of the siderophore periplasmic binding protein FtsB mutant Y137A from Streptococcus pyogenes with ferrioxamine E bound | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, FE (III) ION, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XFI
 
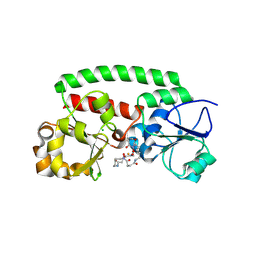 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with ferrioxamine E bound (crystal form 2) | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, CHLORIDE ION, FE (III) ION, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XET
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XF4
 
 | | Structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with Bisucaberin bound | | Descriptor: | 1,12-bis(oxidanyl)-1,6,12,17-tetrazacyclodocosane-2,5,13,16-tetrone, FE (III) ION, Iron-hydroxamate ABC transporter substrate-binding protein FtsB | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XF8
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with ferrioxamine B | | Descriptor: | 1,2-ETHANEDIOL, Ferrioxamine B, Iron-hydroxamate ABC transporter substrate-binding protein FtsB, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XF9
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB mutant Y137A from Streptococcus pyogenes | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
7POO
 
 | | Crystal structure of profragilysin-3 (proBFT-3) from Bacteroides fragilis in complex with foliosidine in P212121. | | Descriptor: | ACETATE ION, BFT-3, PROLINE, ... | | Authors: | Eckhard, U, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis.
Protein Sci., 31, 2022
|
|
7POQ
 
 | | Crystal structure of profragilysin-3 (proBFT-3) from Bacteroides fragilis in complex with foliosidine in P41212. | | Descriptor: | 1,2-ETHANEDIOL, BFT-3, CHLORIDE ION, ... | | Authors: | Eckhard, U, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis.
Protein Sci., 31, 2022
|
|
7PND
 
 | | Crystal structure of profragilysin-3 (proBFT-3) from Bacteroides fragilis at 1.85 A resolution. | | Descriptor: | BFT-3, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Eckhard, U, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-09-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis.
Protein Sci., 31, 2022
|
|
7POL
 
 | | Crystal structure of profragilysin-3 (proBFT-3) from Bacteroides fragilis in complex with flumequine | | Descriptor: | (12~{R})-7-fluoranyl-12-methyl-4-oxidanylidene-1-azatricyclo[7.3.1.0^{5,13}]trideca-2,5(13),6,8-tetraene-3-carboxylic acid, BFT-3, CHLORIDE ION, ... | | Authors: | Eckhard, U, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis.
Protein Sci., 31, 2022
|
|
7POU
 
 | | Crystal structure of profragilysin-3 (proBFT-3) from Bacteroides fragilis in complex with hesperetin. | | Descriptor: | (2S)-5,7-dihydroxy-2-(3-hydroxy-4-methoxyphenyl)-2,3-dihydro-4H-1-benzopyran-4-one, BFT-3, DIMETHYL SULFOXIDE, ... | | Authors: | Eckhard, U, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis.
Protein Sci., 31, 2022
|
|
