7OQ1
 
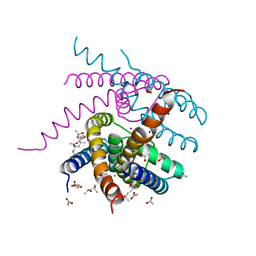 | | NaK S-ELM mutant with Na+ and K+ | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
7OQ2
 
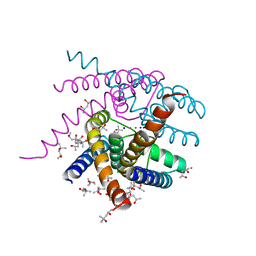 | | NaK S-DI mutant soaked in Na+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
7OUI
 
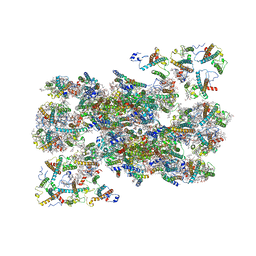 | | Structure of C2S2M2-type Photosystem supercomplex from Arabidopsis thaliana (digitonin-extracted) | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Graca, A.T, Hall, M, Persson, K, Schroder, W.P. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | High-resolution model of Arabidopsis Photosystem II reveals the structural consequences of digitonin-extraction.
Sci Rep, 11, 2021
|
|
7PA0
 
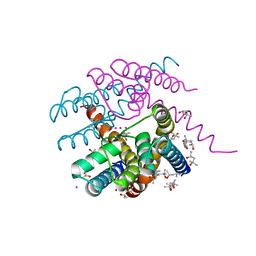 | | NaK C-DI F92A mutant with Rb+ and K+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
6E5G
 
 | |
6GER
 
 | |
6TYG
 
 | | Crystal structure of MTB sigma L transcription initiation complex with 9 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*GP*GP*TP*G)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6U1V
 
 | | Crystal structure of acyl-ACP/acyl-CoA dehydrogenase from allylmalonyl-CoA and FK506 biosynthesis, TcsD | | Descriptor: | Acyl-CoA dehydrogenase domain-containing protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Blake-Hedges, J.M, Pereira, J.H, Barajas, J.F, Adams, P.D, Keasling, J.D. | | Deposit date: | 2019-08-16 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Mechanism of Regioselectivity in an Unusual Bacterial Acyl-CoA Dehydrogenase.
J.Am.Chem.Soc., 142, 2020
|
|
6U0O
 
 | | Crystal structure of a peptidoglycan release complex, SagB-SpdC, in lipidic cubic phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(2-ETHOXYETHOXY)ETHANOL, CITRATE ANION, ... | | Authors: | Owens, T.W, Schaefer, K, Kahne, D, Walker, S. | | Deposit date: | 2019-08-14 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and reconstitution of a hydrolase complex that may release peptidoglycan from the membrane after polymerization.
Nat Microbiol, 6, 2021
|
|
6TX7
 
 | |
6TYE
 
 | | Crystal structure of MTB sigma L transcription initiation complex with 5 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ULX
 
 | |
6TX5
 
 | |
6VBK
 
 | | Crystal structure of N-terminal domain of Mycobacterium tuberculosis complex Lon protease | | Descriptor: | GLYCEROL, Lon211 | | Authors: | Bi, F.K, Chen, C, Chen, X.Y, Guo, C.Y, Lin, D.H. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the N domain of Lon protease from Mycobacterium avium complex.
Protein Sci., 28, 2019
|
|
6UP0
 
 | |
6RPQ
 
 | | Crystal structure of PhoCDC21-1 intein | | Descriptor: | Ubiquitin-like protein SMT3,1108aa long hypothetical cell division control protein | | Authors: | Beyer, H.M, Mikula, K.M, Iwai, H. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Crystal structures of CDC21-1 inteins from hyperthermophilic archaea reveal the selection mechanism for the highly conserved homing endonuclease insertion site.
Extremophiles, 23, 2019
|
|
6RPP
 
 | | Crystal structure of PabCDC21-1 intein | | Descriptor: | ACETATE ION, Cell division control protein, DI(HYDROXYETHYL)ETHER | | Authors: | Mikula, K.M, Beyer, H.M, Iwai, H. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of CDC21-1 inteins from hyperthermophilic archaea reveal the selection mechanism for the highly conserved homing endonuclease insertion site.
Extremophiles, 23, 2019
|
|
6SH7
 
 | |
6WYH
 
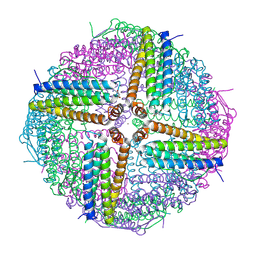 | |
6WYG
 
 | |
6TDB
 
 | | Neuropilin2-b1 domain in a complex with the C-terminal VEGFB167 peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, C-terminal VEGFB167 peptide, ... | | Authors: | Eldrid, C, Yu, L, Yelland, T, Fotinou, C, Djordjevic, S. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | NRP2-b1 domain in a complex with the C-terminal VEGFB167 peptide
To Be Published
|
|
6WYF
 
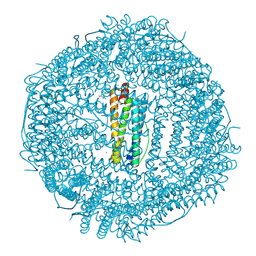 | |
6WPM
 
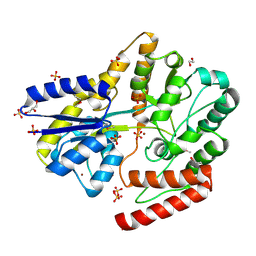 | | Crystal structure of a putative oligosaccharide periplasmic-binding protein from Synechococcus sp. MITs9220 in complex with zinc | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Ford, B.A, Michie, K.A, Paulsen, I.T, Mabbutt, B.C, Shah, B.S. | | Deposit date: | 2020-04-27 | | Release date: | 2021-05-12 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Novel functional insights into a modified sugar-binding protein from Synechococcus MITS9220.
Sci Rep, 12, 2022
|
|
6TX4
 
 | |
6TYF
 
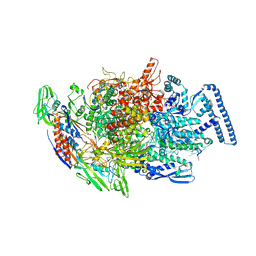 | | Crystal structure of MTB sigma L transcription initiation complex with 6 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*GP*GP*T)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
