6CS1
 
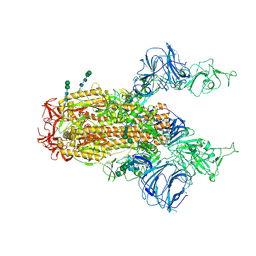 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, two S1 CTDs in an upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CRX
 
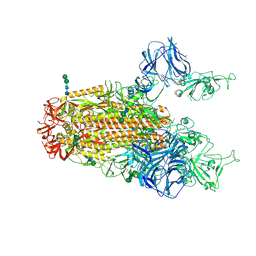 | | SARS Spike Glycoprotein, Stabilized variant, two S1 CTDs in the upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CRW
 
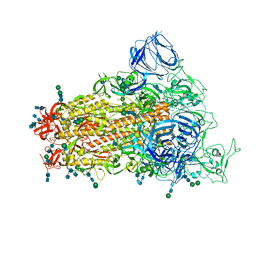 | | SARS Spike Glycoprotein, Stabilized variant, single upwards S1 CTD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
8EQA
 
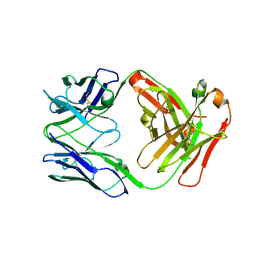 | | Crystal structure of human anti-N1 neuraminidase 2H08 Fab | | Descriptor: | 2H08 Fab heavy chain, 2H08 Fab light chain, ZINC ION | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2022-10-07 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Human anti-N1 monoclonal antibodies elicited by pandemic H1N1 virus infection broadly inhibit HxN1 viruses in vitro and in vivo.
Immunity, 56, 2023
|
|
8EQC
 
 | | Crystal structure of human anti-N1 neuraminidase 3H03 Fab | | Descriptor: | 3H03 Fab heavy chain, 3H03 Fab light chain, PHOSPHATE ION | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2022-10-07 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human anti-N1 monoclonal antibodies elicited by pandemic H1N1 virus infection broadly inhibit HxN1 viruses in vitro and in vivo.
Immunity, 56, 2023
|
|
7U04
 
 | | IOMA class antibody ACS101 | | Descriptor: | GLYCEROL, IOMA class antibody ACS101 heavy chain, IOMA class antibody ACS101 light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7U0K
 
 | | IOMA class antibody Fab ACS124 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, IOMA Class antibody ACS124 Heavy chain, IOMA Class antibody ACS124 Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7U5G
 
 | | ACS122 Fab | | Descriptor: | ACS122 Fab Heavy chain, ACS122 Fab Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-03-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
7Z3A
 
 | | AMC009 SOSIPv5.2 in complex with Fabs ACS101 and ACS124 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACS101 heavy chain, ... | | Authors: | van Schooten, J, Ward, A. | | Deposit date: | 2022-03-02 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7ZLK
 
 | | AMC009 SOSIPv5.2 in complex with Fabs ACS101 and ACS124 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACS114 heavy chain, ... | | Authors: | van Schooten, J, Ozorowski, G, Ward, A. | | Deposit date: | 2022-04-15 | | Release date: | 2022-09-21 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
7JN5
 
 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 heavy chain, ... | | Authors: | Wu, N.C, Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | A natural mutation between SARS-CoV-2 and SARS-CoV determines neutralization by a cross-reactive antibody.
Plos Pathog., 16, 2020
|
|
8D0Z
 
 | | S728-1157 IgG in complex with SARS-CoV-2-6P-Mut7 Spike protein (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S728-1157 Fab heavy chain variable region, S728-1157 Fab light chain variable region, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2022-05-26 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Site of vulnerability on SARS-CoV-2 spike induces broadly protective antibody against antigenically distinct Omicron subvariants.
J.Clin.Invest., 133, 2023
|
|
8ELP
 
 | |
8ELO
 
 | |
8ELQ
 
 | |
6U59
 
 | | HIV-1 B41 SOSIP.664 in complex with rabbit antibody 13B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SOSIP.664 gp120,SOSIP.664 gp120, ... | | Authors: | Yang, Y.R, Ward, A.B. | | Deposit date: | 2019-08-27 | | Release date: | 2020-01-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Autologous Antibody Responses to an HIV Envelope Glycan Hole Are Not Easily Broadened in Rabbits.
J.Virol., 94, 2020
|
|
6UIG
 
 | |
6UYE
 
 | | EBOV GPdMuc Makona bound to rEBOV-548 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody rEBOV-548 Fab, ... | | Authors: | Murin, C.D, Ward, A.B. | | Deposit date: | 2019-11-13 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Analysis of a Therapeutic Antibody Cocktail Reveals Determinants for Cooperative and Broad Ebolavirus Neutralization.
Immunity, 52, 2020
|
|
6VFK
 
 | |
6VFL
 
 | |
6V0R
 
 | | BG505 SOSIP.664 Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nogal, B, Cottrell, C.A, Ward, A.B. | | Deposit date: | 2019-11-19 | | Release date: | 2020-04-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Mapping Polyclonal Antibody Responses in Non-human Primates Vaccinated with HIV Env Trimer Subunit Vaccines.
Cell Rep, 30, 2020
|
|
6VO0
 
 | | BG505 SOSIP.v5.2 in complex with rabbit Fab 43A2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 43A2 heavy chain, ... | | Authors: | Nogal, B, Cottrell, C.A, Ward, A.B. | | Deposit date: | 2020-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | HIV envelope trimer-elicited autologous neutralizing antibodies bind a region overlapping the N332 glycan supersite.
Sci Adv, 6, 2020
|
|
5UMN
 
 | |
5VZR
 
 | |
5VYH
 
 | | Crystal Structure of MERS-CoV S1 N-terminal Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Wang, N, Wrapp, D, Pallesen, J, Ward, A.B, McLellan, J.S. | | Deposit date: | 2017-05-25 | | Release date: | 2017-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
