7NTU
 
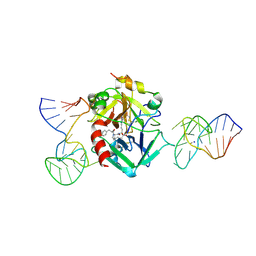 | | X-ray structure of the complex between human alpha thrombin and two duplex/quadruplex aptamers: NU172 and HD22_27mer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD22_27mer, ... | | Authors: | Troisi, R, Santamaria, A, Sica, F. | | Deposit date: | 2021-03-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and functional analysis of the simultaneous binding of two duplex/quadruplex aptamers to human alpha-thrombin.
Int.J.Biol.Macromol., 181, 2021
|
|
6EVV
 
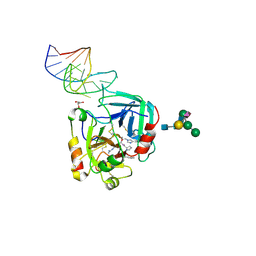 | | X-ray structure of the complex between human alpha thrombin and NU172, a duplex/quadruplex 26-mer DNA aptamer, in the presence of potassium ions. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Troisi, R, Russo Krauss, I, Sica, F. | | Deposit date: | 2017-11-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer.
Nucleic Acids Res., 46, 2018
|
|
6GN7
 
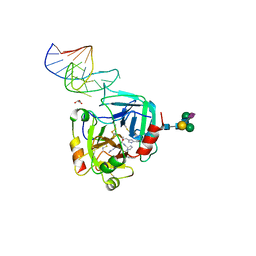 | | X-ray structure of the complex between human alpha thrombin and NU172, a duplex/quadruplex 26-mer DNA aptamer, in the presence of sodium ions. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Troisi, R, Russo Krauss, I, Sica, F. | | Deposit date: | 2018-05-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer.
Nucleic Acids Res., 46, 2018
|
|
7ZKN
 
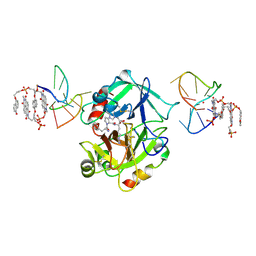 | | X-ray structure of the complex between human alpha thrombin and a pseudo-cyclic thrombin binding aptamer (TBA-NNp/DDp) - Crystal form gamma | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[13-methyl-5,7,12,14-tetrakis(oxidanylidene)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1(15),2,4(16),8,10-pentaen-6-yl]propyl 3-[5,7,12,14-tetrakis(oxidanylidene)-13-(3-oxidanylpropyl)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1,3,8(16),9,11(15)-pentaen-6-yl]propyl hydrogen phosphate, 3-[5-[3-bis(oxidanyl)phosphanyloxypropoxy]naphthalen-1-yl]oxypropyl 3-(5-oxidanylnaphthalen-1-yl)oxypropyl hydrogen phosphate, ... | | Authors: | Troisi, R, Sica, F. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | A terminal functionalization strategy reveals unusual binding abilities of anti-thrombin anticoagulant aptamers.
Mol Ther Nucleic Acids, 30, 2022
|
|
7ZKL
 
 | | X-ray structure of the complex between human alpha thrombin and a pseudo-cyclic thrombin binding aptamer (TBA-NNp/DDp) - Crystal form alpha | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 3-[13-methyl-5,7,12,14-tetrakis(oxidanylidene)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1(15),2,4(16),8,10-pentaen-6-yl]propyl 3-[5,7,12,14-tetrakis(oxidanylidene)-13-(3-oxidanylpropyl)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1,3,8(16),9,11(15)-pentaen-6-yl]propyl hydrogen phosphate, 3-[5-[3-bis(oxidanyl)phosphanyloxypropoxy]naphthalen-1-yl]oxypropyl 3-(5-oxidanylnaphthalen-1-yl)oxypropyl hydrogen phosphate, ... | | Authors: | Troisi, R, Sica, F. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | A terminal functionalization strategy reveals unusual binding abilities of anti-thrombin anticoagulant aptamers.
Mol Ther Nucleic Acids, 30, 2022
|
|
7ZKO
 
 | | X-ray structure of the complex between human alpha thrombin and a pseudo-cyclic thrombin binding aptamer (TBA-NNp/DDp) - Crystal form delta | | Descriptor: | 3-[13-methyl-5,7,12,14-tetrakis(oxidanylidene)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1(15),2,4(16),8,10-pentaen-6-yl]propyl 3-[5,7,12,14-tetrakis(oxidanylidene)-13-(3-oxidanylpropyl)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1,3,8(16),9,11(15)-pentaen-6-yl]propyl hydrogen phosphate, 3-[5-[3-bis(oxidanyl)phosphanyloxypropoxy]naphthalen-1-yl]oxypropyl 3-(5-oxidanylnaphthalen-1-yl)oxypropyl hydrogen phosphate, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Troisi, R, Sica, F. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A terminal functionalization strategy reveals unusual binding abilities of anti-thrombin anticoagulant aptamers.
Mol Ther Nucleic Acids, 30, 2022
|
|
7ZKM
 
 | | X-ray structure of the complex between human alpha thrombin and a pseudo-cyclic thrombin binding aptamer (TBA-NNp/DDp) - Crystal form beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[13-methyl-5,7,12,14-tetrakis(oxidanylidene)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1(15),2,4(16),8,10-pentaen-6-yl]propyl 3-[5,7,12,14-tetrakis(oxidanylidene)-13-(3-oxidanylpropyl)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1,3,8(16),9,11(15)-pentaen-6-yl]propyl hydrogen phosphate, 3-[5-[3-bis(oxidanyl)phosphanyloxypropoxy]naphthalen-1-yl]oxypropyl 3-(5-oxidanylnaphthalen-1-yl)oxypropyl hydrogen phosphate, ... | | Authors: | Troisi, R, Sica, F. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A terminal functionalization strategy reveals unusual binding abilities of anti-thrombin anticoagulant aptamers.
Mol Ther Nucleic Acids, 30, 2022
|
|
8BRC
 
 | | Crystal structure of the adduct between human serum transferrin and cisplatin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIA, FE (III) ION, ... | | Authors: | Troisi, R, Galardo, F, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2022-11-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Cisplatin Binding to Human Serum Transferrin: A Crystallographic Study.
Inorg.Chem., 62, 2023
|
|
8BW5
 
 | | X-ray structure of the complex between human alpha thrombin and the duplex/quadruplex aptamer M08s-1_41mer | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, M08s-1_41mer, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Troisi, R, Napolitano, V, Sica, F. | | Deposit date: | 2022-12-06 | | Release date: | 2023-07-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Steric hindrance and structural flexibility shape the functional properties of a guanine-rich oligonucleotide.
Nucleic Acids Res., 51, 2023
|
|
8C64
 
 | | Crystal structure of arsenoplatin-1/B-DNA adduct obtained upon 48 h of soaking | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PLATINUM (II) ION, arsenoplatin-1 | | Authors: | Troisi, R, Tito, G, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2023-01-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | On the mechanism of action of arsenoplatins: arsenoplatin-1 binding to a B-DNA dodecamer.
Dalton Trans, 53, 2024
|
|
8C62
 
 | | Crystal structure of cisplatin/B-DNA adduct | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PLATINUM (II) ION | | Authors: | Troisi, R, Tito, G, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2023-01-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | On the mechanism of action of arsenoplatins: arsenoplatin-1 binding to a B-DNA dodecamer.
Dalton Trans, 53, 2024
|
|
8C63
 
 | | Crystal structure of arsenoplatin-1/B-DNA adduct obtained upon 4 h of soaking | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, PLATINUM (II) ION, ... | | Authors: | Troisi, R, Tito, G, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2023-01-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | On the mechanism of action of arsenoplatins: arsenoplatin-1 binding to a B-DNA dodecamer.
Dalton Trans, 53, 2024
|
|
8RI5
 
 | | Crystal structure of transplatin/B-DNA adduct obtained upon 48 h of soaking | | Descriptor: | AMMONIA, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Tito, G, Troisi, R, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2023-12-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.415 Å) | | Cite: | On the mechanism of action of arsenoplatins: arsenoplatin-1 binding to a B-DNA dodecamer.
Dalton Trans, 53, 2024
|
|
8RI3
 
 | | Crystal structure of transplatin/B-DNA adduct obtained upon 7 days of soaking | | Descriptor: | AMMONIA, CHLORIDE ION, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), ... | | Authors: | Tito, G, Troisi, R, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2023-12-18 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | On the mechanism of action of arsenoplatins: arsenoplatin-1 binding to a B-DNA dodecamer.
Dalton Trans, 53, 2024
|
|
6Z8W
 
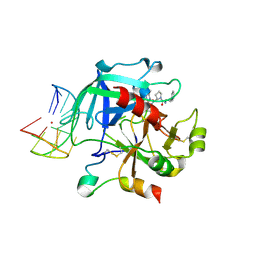 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3G), which contains 1-beta-D-glucopyranosyl residue in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
6Z8X
 
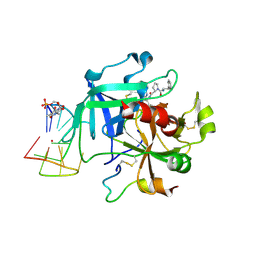 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3Leu), which contains leucyl amide in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
6Z8V
 
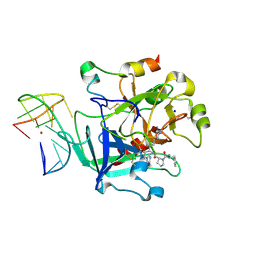 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3L), which contains 1-beta-D-lactopyranosyl residue in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
7BFL
 
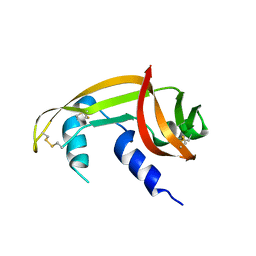 | | X-ray structure of SS-RNase-2 des116-120 | | Descriptor: | Angiogenin-1 | | Authors: | Sica, F, Russo Krauss, I, Troisi, R. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | The structural features of an ancient ribonuclease from Salmo salar reveal an intriguing case of auto-inhibition.
Int.J.Biol.Macromol., 182, 2021
|
|
7BFK
 
 | | X-ray structure of SS-RNase-2 | | Descriptor: | Angiogenin-1 | | Authors: | Sica, F, Russo Krauss, I, Troisi, R. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The structural features of an ancient ribonuclease from Salmo salar reveal an intriguing case of auto-inhibition.
Int.J.Biol.Macromol., 182, 2021
|
|
8CE2
 
 | | X-ray structure of the adduct formed upon reaction of a B-DNA double helical dodecamer with dirhodium tetraacetate | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Tito, G, Troisi, R, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2023-02-01 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Dirhodium tetraacetate binding to a B-DNA double helical dodecamer probed by X-ray crystallography and mass spectrometry.
Dalton Trans, 52, 2023
|
|
6SVF
 
 | | Crystal structure of the P235GK mutant of ArgBP from T. maritima | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein | | Authors: | Vitagliano, L, Berisio, R, Esposito, L, Balasco, N, Smaldone, G, Ruggiero, A. | | Deposit date: | 2019-09-18 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The non-swapped monomeric structure of the arginine-binding protein from Thermotoga maritima.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
