1SML
 
 | | METALLO BETA LACTAMASE L1 FROM STENOTROPHOMONAS MALTOPHILIA | | Descriptor: | PROTEIN (PENICILLINASE), ZINC ION | | Authors: | Ullah, J.H, Walsh, T.R, Taylor, I.A, Emery, D.C, Verma, C.S, Gamblin, S.J, Spencer, J. | | Deposit date: | 1998-09-22 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia at 1.7 A resolution.
J.Mol.Biol., 284, 1998
|
|
1SW6
 
 | |
1U7K
 
 | | Structure of a hexameric N-terminal domain from murine leukemia virus capsid | | Descriptor: | Gag polyprotein | | Authors: | Mortuza, G.B, Haire, L.F, Stevens, A, Smerdon, S.J, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-resolution structure of a retroviral capsid hexameric amino-terminal domain.
Nature, 431, 2004
|
|
2X1A
 
 | | Structure of Rna15 RRM with RNA bound (G) | | Descriptor: | 5'-R(*GP*UP*UP*GP*UP)-3', MAGNESIUM ION, MRNA 3'-END-PROCESSING PROTEIN RNA15 | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2X1F
 
 | | Structure of Rna15 RRM with bound RNA (GU) | | Descriptor: | 5'-R(*GP*UP*UP*GP*UP)-3', MRNA 3'-END-PROCESSING PROTEIN RNA15 | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2X1B
 
 | | Structure of RNA15 RRM | | Descriptor: | MRNA 3'-END-PROCESSING PROTEIN RNA15, PHOSPHATE ION | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2Y4Z
 
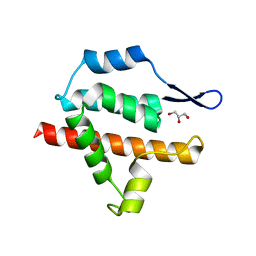 | | Structure of the amino-terminal capsid restriction escape mutation N- MLV L10W | | Descriptor: | CAPSID PROTEIN P30, GLYCEROL | | Authors: | Goldstone, D.C, Holden-Dye, K, Ohkura, S, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2011-01-11 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Novel Escape Mutants Suggest an Extensive Trim5Alpha Binding Site Spanning the Entire Outer Surface of the Murine Leukemia Virus Capsid Protein.
Plos Pathog., 7, 2011
|
|
1L3G
 
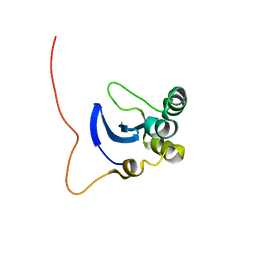 | | NMR Structure of the DNA-binding Domain of Cell Cycle Protein, Mbp1(2-124) from Saccharomyces cerevisiae | | Descriptor: | TRANSCRIPTION FACTOR Mbp1 | | Authors: | Nair, M, McIntosh, P.B, Frenkiel, T.A, Kelly, G, Taylor, I.A, Smerdon, S.J, Lane, A.N. | | Deposit date: | 2002-02-27 | | Release date: | 2003-02-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the DNA-Binding Domain of the Cell Cycle Protein Mbp1 from Saccharomyces cerevisiae
Biochemistry, 42, 2003
|
|
3U1N
 
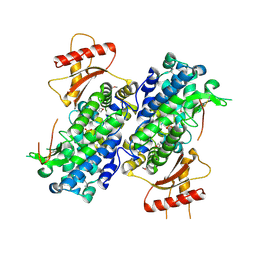 | | Structure of the catalytic core of human SAMHD1 | | Descriptor: | PHOSPHATE ION, SAM domain and HD domain-containing protein 1, ZINC ION | | Authors: | Goldstone, D.C, Ennis-Adeniran, V, Walker, P.A, Haire, L.F, Webb, M, Taylor, I.A. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase
Nature, 480, 2011
|
|
1O9S
 
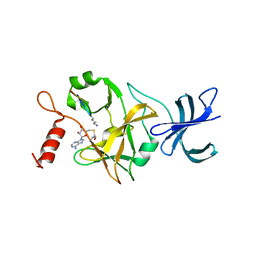 | | Crystal structure of a ternary complex of the human histone methyltransferase SET7/9 | | Descriptor: | GENE FRAGMENT FOR HISTONE H3, HISTONE-LYSINE N-METHYLTRANSFERASE, H3 LYSINE-4 SPECIFIC, ... | | Authors: | Xiao, B, Jing, C, Wilson, J.R, Walker, P.A, Vasisht, N, Kelly, G, Howell, S, Taylor, I.A, Blackburn, G.M, Gamblin, S.J. | | Deposit date: | 2002-12-18 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Catalytic Mechanism of the Human Histone Methyltransferase Set7/9
Nature, 421, 2003
|
|
8QHQ
 
 | | Crystal structure of human DNPH1 bound to hmdUMP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE | | Authors: | Rzechorzek, N.J, West, S.C. | | Deposit date: | 2023-09-09 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism of substrate hydrolysis by the human nucleotide pool sanitiser DNPH1.
Nat Commun, 14, 2023
|
|
8QHR
 
 | | Crystal structure of the human DNPH1 glycosyl-enzyme intermediate | | Descriptor: | 1',2'-DIDEOXYRIBOFURANOSE-5'-PHOSPHATE, 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, ... | | Authors: | Rzechorzek, N.J, West, S.C. | | Deposit date: | 2023-09-09 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of substrate hydrolysis by the human nucleotide pool sanitiser DNPH1.
Nat Commun, 14, 2023
|
|
4USG
 
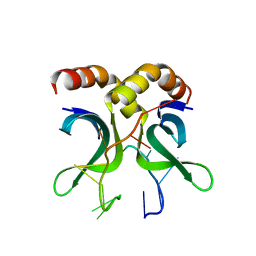 | | Crystal structure of PC4 W89Y mutant complex with DNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP *TP*TP*TP*TP*TP*G)-3', ACTIVATED RNA POLYMERASE II TRANSCRIPTIONAL COACTIVATOR P15 | | Authors: | Zhao, Y, Liu, J. | | Deposit date: | 2014-07-08 | | Release date: | 2015-03-18 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Substitution of Tryptophan 89 with Tyrosine Switches the DNA Binding Mode of Pc4.
Sci.Rep., 5, 2015
|
|
7ZPP
 
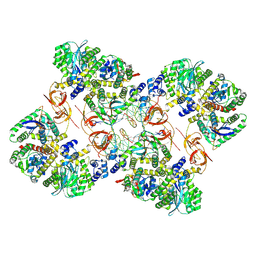 | | Cryo-EM structure of the MVV CSC intasome at 4.5A resolution | | Descriptor: | Integrase, vDNA, non-transferred strand, ... | | Authors: | Ballandras-Colas, A, Maskell, D, Pye, V.E, Locke, J, Swuec, S, Kotecha, A, Costa, A, Cherepanov, P. | | Deposit date: | 2022-04-28 | | Release date: | 2022-05-11 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A supramolecular assembly mediates lentiviral DNA integration
Science, 355, 2017
|
|
7Z1Z
 
 | | MVV strand transfer complex (STC) intasome in complex with LEDGF/p75 at 3.5 A resolution | | Descriptor: | DNA (37-MER), DNA (5'-D(*GP*CP*TP*GP*CP*GP*AP*GP*AP*TP*CP*CP*GP*CP*TP*CP*CP*GP*GP*TP*G)-3'), DNA (5'-D(P*TP*TP*GP*AP*TP*TP*AP*GP*GP*GP*TP*G)-3'), ... | | Authors: | Pye, V.E, Ballandras-Colas, A, Cherepanov, P. | | Deposit date: | 2022-02-25 | | Release date: | 2022-05-11 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Multivalent interactions essential for lentiviral integrase function.
Nat Commun, 13, 2022
|
|
8A1P
 
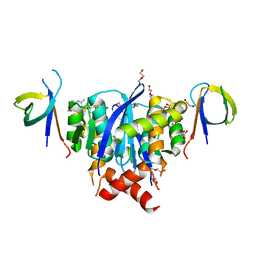 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor BI-D | | Descriptor: | (2S)-tert-butoxy[4-(3,4-dihydro-2H-chromen-6-yl)-2-methylquinolin-3-yl]ethanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, M.R, Pye, V.E, Cook, N.J, Cherepanov, P. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Drug-Induced Interface That Drives HIV-1 Integrase Hypermultimerization and Loss of Function.
Mbio, 14, 2023
|
|
8A1Q
 
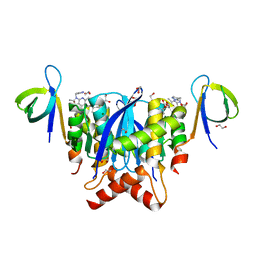 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor STP0404 (Pirmitegravir) | | Descriptor: | (2S)-tert-butoxy{4-(4-chlorophenyl)-2,3,6-trimethyl-1-[(1-methyl-1H-pyrazol-4-yl)methyl]-1H-pyrrolo[2,3-b]pyridin-5-yl}acetic acid, 1,2-ETHANEDIOL, Integrase, ... | | Authors: | Singer, M.R, Pye, V.E, Cook, N.J, Cherepanov, P. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The Drug-Induced Interface That Drives HIV-1 Integrase Hypermultimerization and Loss of Function.
Mbio, 14, 2023
|
|
8RIL
 
 | |
8RJ3
 
 | |
8RJW
 
 | | Human RAD52 open ring - ssDNA complex | | Descriptor: | DNA repair protein RAD52 homolog, MAGNESIUM ION, ssDNA | | Authors: | Liang, C.C, West, S.C. | | Deposit date: | 2023-12-21 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Mechanism of single-stranded DNA annealing by RAD52-RPA complex
Nature, 2024
|
|
8RK2
 
 | |
6ZX9
 
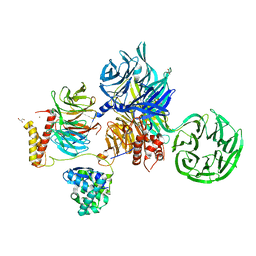 | | Crystal structure of SIV Vpr,fused to T4 lysozyme, isolated from moustached monkey, bound to human DDB1 and human DCAF1 (amino acid residues 1046-1396) | | Descriptor: | DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, GLYCEROL, ... | | Authors: | Schwefel, D, Banchenko, S. | | Deposit date: | 2020-07-29 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.519729 Å) | | Cite: | Structural insights into Cullin4-RING ubiquitin ligase remodelling by Vpr from simian immunodeficiency viruses.
Plos Pathog., 17, 2021
|
|
6IR8
 
 | | Rice WRKY/DNA complex | | Descriptor: | DNA (5'-D(P*GP*AP*TP*AP*TP*TP*TP*GP*AP*CP*CP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*GP*GP*TP*CP*AP*AP*AP*TP*AP*TP*C)-3'), OsWRKY45, ... | | Authors: | Liu, J, Cheng, X, Wang, D. | | Deposit date: | 2018-11-12 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of dimerization and dual W-box DNA recognition by rice WRKY domain.
Nucleic Acids Res., 47, 2019
|
|
5HXL
 
 | |
5HYC
 
 | | Structure based function annotation of a hypothetical protein MGG_01005 related to the development of rice blast fungus | | Descriptor: | Cytoplasmic dynein 1 intermediate chain 2, Uncharacterized protein | | Authors: | Liu, J, Li, G, Huang, J, Peng, Y.-l. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure based function-annotation of hypothetical protein MGG_01005 from Magnaporthe oryzae reveals it is the dynein light chain orthologue of dynlt1/3.
Sci Rep, 8, 2018
|
|
