2H89
 
 | | Avian Respiratory Complex II with Malonate Bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Huang, L.S, Shen, J.T, Wang, A.C, Berry, E.A. | | Deposit date: | 2006-06-06 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the
Biochim.Biophys.Acta, 1757
|
|
2H88
 
 | | Avian Mitochondrial Respiratory Complex II at 1.8 Angstrom Resolution | | Descriptor: | AZIDE ION, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Huang, L.S, Shen, J.T, Wang, A.C, Berry, E.A. | | Deposit date: | 2006-06-06 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the
Biochim.Biophys.Acta, 1757
|
|
4Z55
 
 | |
3K5V
 
 | | Structure of Abl kinase in complex with imatinib and GNF-2 | | Descriptor: | 3-(6-{[4-(trifluoromethoxy)phenyl]amino}pyrimidin-4-yl)benzamide, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, CHLORIDE ION, ... | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Rummel, G, Strauss, A. | | Deposit date: | 2009-10-08 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors.
Nature, 463, 2010
|
|
5AZC
 
 | |
5AZB
 
 | | Crystal structure of Escherichia coli Lgt in complex with phosphatidylglycerol and the inhibitor palmitic acid | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PALMITIC ACID, Prolipoprotein diacylglyceryl transferase, ... | | Authors: | Zhang, X.C, Mao, G, Zhao, Y. | | Deposit date: | 2015-09-30 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of E. coli lipoprotein diacylglyceryl transferase
Nat Commun, 7, 2016
|
|
2F9Y
 
 | | The Crystal Structure of The Carboxyltransferase Subunit of ACC from Escherichia coli | | Descriptor: | Acetyl-CoA carboxylase, Carboxyltransferase alpha chain, Acetyl-coenzyme A carboxylase carboxyl transferase subunit beta, ... | | Authors: | Bilder, P.W. | | Deposit date: | 2005-12-06 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Structure of the Carboxyltransferase Component of Acetyl-CoA Carboxylase Reveals a Zinc-Binding Motif Unique to the Bacterial Enzyme(,).
Biochemistry, 45, 2006
|
|
2ESW
 
 | |
2F9I
 
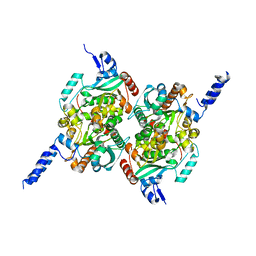 | | Crystal Structure of the carboxyltransferase subunit of ACC from Staphylococcus aureus | | Descriptor: | ZINC ION, acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, acetyl-coenzyme A carboxylase carboxyl transferase subunit beta | | Authors: | Bilder, P.W. | | Deposit date: | 2005-12-05 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The structure of the carboxyltransferase component of acetyl-coA carboxylase reveals a zinc-binding motif unique to the bacterial enzyme.
Biochemistry, 45, 2006
|
|
3T8R
 
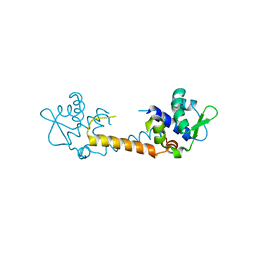 | | Crystal structure of Staphylococcus aureus CymR | | Descriptor: | Staphylococcus aureus CymR | | Authors: | He, C, Ji, Q. | | Deposit date: | 2011-08-01 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Staphylococcus aureus CymR Is a New Thiol-based Oxidation-sensing Regulator of Stress Resistance and Oxidative Response.
J.Biol.Chem., 287, 2012
|
|
3T8T
 
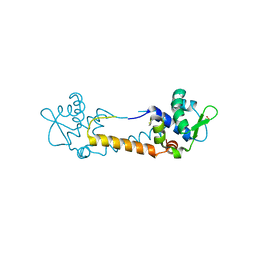 | | Crystal structure of Staphylococcus aureus CymR oxidized form | | Descriptor: | Staphylococcus aureus CymR (oxidized form) | | Authors: | He, C, Ji, Q. | | Deposit date: | 2011-08-01 | | Release date: | 2012-05-16 | | Last modified: | 2012-07-18 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Staphylococcus aureus CymR Is a New Thiol-based Oxidation-sensing Regulator of Stress Resistance and Oxidative Response.
J.Biol.Chem., 287, 2012
|
|
5XHQ
 
 | | Apolipoprotein N-acyl Transferase | | Descriptor: | Apolipoprotein N-acyltransferase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yingzhi, X, Yong, X, Guangyuan, L, Fei, S. | | Deposit date: | 2017-04-23 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | Crystal structure of E. coli apolipoprotein N-acyl transferase
Nat Commun, 8, 2017
|
|
4MFH
 
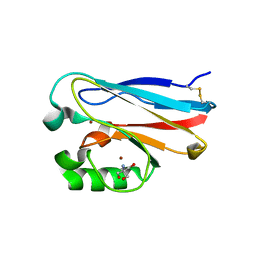 | | Crystal Structure of M121G Azurin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Tian, S, Lu, Y. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-15 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Copper-sulfenate complex from oxidation of a cavity mutant of Pseudomonas aeruginosa azurin.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4HQE
 
 | | The crystal structure of QsrR-DNA complex | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*TP*AP*AP*TP*TP*AP*TP*TP*AP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*AP*TP*AP*AP*TP*TP*AP*TP*AP*CP*T)-3'), Transcriptional regulator QsrR | | Authors: | He, C, Ji, Q, Zhang, L. | | Deposit date: | 2012-10-25 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Molecular mechanism of quinone signaling mediated through S-quinonization of a YodB family repressor QsrR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4I88
 
 | | R107G HSP16.5 | | Descriptor: | Small heat shock protein HSP16.5 | | Authors: | Pohl, E, Williamson, I.R, Quinlan, R.A. | | Deposit date: | 2012-12-03 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Changes in the quaternary structure and function of MjHSP16.5 attributable to deletion of the IXI motif and introduction of the substitution, R107G, in the alpha-crystallin domain.
PHILOS.TRANS.R.SOC.LOND.B BIOL.SCI., 368, 2013
|
|
8JT8
 
 | | Crystal structure of 5-HT2AR in complex with (R)-IHCH-7179 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Chen, Z, Fan, L, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 2024
|
|
8JT6
 
 | | 5-HT1A-Gi in complex with compound (R)-IHCH-7179 | | Descriptor: | 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Chen, Z, Xu, P, Huang, S, Xu, H.E, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 2024
|
|
7EPF
 
 | | Crystal structure of mGlu2 bound to NAM597 | | Descriptor: | (8~{R})-4-[2,4-bis(fluoranyl)phenyl]-8-methyl-7-[(2-methylpyrazol-3-yl)methyl]-6,8-dihydro-5~{H}-1,7-naphthyridine-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPE
 
 | | Crystal structure of mGlu2 bound to NAM563 | | Descriptor: | 4-(1-methylpyrazol-4-yl)-7-[[(2~{S})-2-(trifluoromethyl)morpholin-4-yl]methyl]quinoline-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
6KLS
 
 | | Hyperthermophilic respiratory Complex III | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, Cytochrome c, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guohong, P, Guoliang, Z, Hui, Z, Shuangbo, Z, Xiaoyun, P, Yan, Z. | | Deposit date: | 2019-07-30 | | Release date: | 2020-05-13 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A 3.3 angstrom -Resolution Structure of Hyperthermophilic Respiratory Complex III Reveals the Mechanism of Its Thermal Stability.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6KLV
 
 | | Hyperthermophilic respiratory Complex III | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, ANTIMYCIN, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guohong, P, Guoliang, Z, Hui, Z, Shuangbo, Z, Xiaoyun, P, Yan, Z. | | Deposit date: | 2019-07-30 | | Release date: | 2020-05-13 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A 3.3 angstrom -Resolution Structure of Hyperthermophilic Respiratory Complex III Reveals the Mechanism of Its Thermal Stability.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7DEG
 
 | | Cryo-EM structure of a heme-copper terminal oxidase dimer provides insights into its catalytic mechanism | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guoliang, Z, Shuangbo, Z. | | Deposit date: | 2020-11-04 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The Unusual Homodimer of a Heme-Copper Terminal Oxidase Allows Itself to Utilize Two Electron Donors.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6MYR
 
 | | Avian mitochondrial complex II with thiapronil bound | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Huang, L.-S, Luemmen, P, Berry, E.A. | | Deposit date: | 2018-11-02 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6MYS
 
 | | Avian mitochondrial complex II with Atpenin A5 bound, sidechain outside | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 3-[(2S,4S,5R)-5,6-DICHLORO-2,4-DIMETHYL-1-OXOHEXYL]-4-HYDROXY-5,6-DIMETHOXY-2(1H)-PYRIDINONE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6MYQ
 
 | | Avian mitochondrial complex II with ferulenol bound | | Descriptor: | (~{Z})-2-oxidanylbut-2-enedioic acid, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 4-oxidanyl-3-[(2~{E},6~{E})-3,7,11-trimethyldodeca-2,6,10-trienyl]chromen-2-one, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
