5A8G
 
 | | Crystal structure of the wild-type Staphylococcus aureus N- acetylneurminic acid lyase in complex with fluoropyruvate | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Stockwell, J, Daniels, A.D, Windle, C.L, Harman, T, Woodhall, T, Trinh, C.H, Lebel, T, Pearson, A.R, Mulholland, K, Berry, A, Nelson, A. | | Deposit date: | 2015-07-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Evaluation of Fluoropyruvate as Nucleophile in Reactions Catalysed by N-Acetyl Neuraminic Acid Lyase Variants: Scope, Limitations and Stereoselectivity.
Org.Biomol.Chem., 14, 2016
|
|
7KJU
 
 | | Cgi121-tRNA complex | | Descriptor: | MAGNESIUM ION, RNA (75-MER) | | Authors: | Ceccarelli, D.F, Beenstock, J, Wan, L.C.K, Sicheri, F. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | A substrate binding model for the KEOPS tRNA modifying complex.
Nat Commun, 11, 2020
|
|
7KJT
 
 | | KEOPS tRNA modifying sub-complex of archaeal Cgi121 and tRNA | | Descriptor: | RNA (70-MER), Regulatory protein Cgi121 | | Authors: | Ceccarelli, D.F, Beenstock, J, Mao, D.Y.L, Sicheri, F. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | A substrate binding model for the KEOPS tRNA modifying complex.
Nat Commun, 11, 2020
|
|
6GFF
 
 | | Structure of GARP (LRRC32) in complex with latent TGF-beta1 and MHG-8 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat-containing protein 32, MHG-8 Fab heavy chain, ... | | Authors: | Merceron, R, Lienart, S, Vanderaa, C, Colau, D, Stockis, J, Van Der Woning, B, De Haard, H, Saunders, M, Coulie, P.G, Savvides, S.N, Lucas, S. | | Deposit date: | 2018-04-30 | | Release date: | 2018-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of latent TGF-beta 1 presentation and activation by GARP on human regulatory T cells.
Science, 362, 2018
|
|
2OUL
 
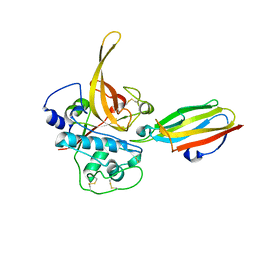 | | The Structure of Chagasin in Complex with a Cysteine Protease Clarifies the Binding Mode and Evolution of a New Inhibitor Family | | Descriptor: | Chagasin, Falcipain 2 | | Authors: | Wang, S.X, Chand, K, Huang, R, Whisstock, J, Jacobelli, J, Fletterick, R.J, Rosenthal, P.J, McKerrow, J.H. | | Deposit date: | 2007-02-11 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of chagasin in complex with a cysteine protease clarifies the binding mode and evolution of an inhibitor family.
Structure, 15, 2007
|
|
2JF7
 
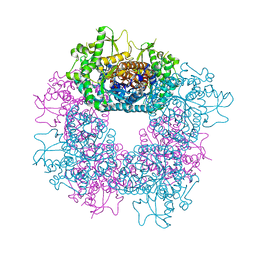 | | Structure of Strictosidine Glucosidase | | Descriptor: | STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Barleben, L, Panjikar, S, Ruppert, M, Koepke, J, Stockigt, J. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Molecular Architecture of Strictosidine Glucosidase - the Gateway to the Biosynthesis of the Monoterpenoid Indole Alkaloid Family
Plant Cell, 19, 2007
|
|
2JF6
 
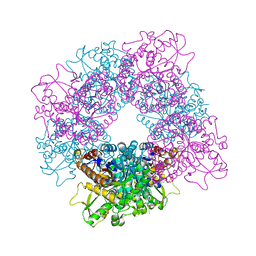 | | Structure of inactive mutant of Strictosidine Glucosidase in complex with strictosidine | | Descriptor: | METHYL (2S,3R,4S)-3-ETHYL-2-(BETA-D-GLUCOPYRANOSYLOXY)-4-[(1S)-2,3,4,9-TETRAHYDRO-1H-BETA-CARBOLIN-1-YLMETHYL]-3,4-DIHYDRO-2H-PYRAN-5-CARBOXYLATE, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Barleben, L, Panjikar, S, Ruppert, M, Koepke, J, Stockigt, J. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Molecular Architecture of Strictosidine Glucosidase - the Gateway to the Biosynthesis of the Monoterpenoid Indole Alkaloid Family
Plant Cell, 19, 2007
|
|
1CHD
 
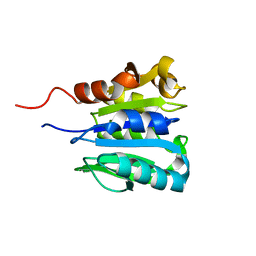 | | CHEB METHYLESTERASE DOMAIN | | Descriptor: | CHEB METHYLESTERASE | | Authors: | West, A.H, Martinez-Hackert, E, Stock, A.M. | | Deposit date: | 1995-03-09 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the catalytic domain of the chemotaxis receptor methylesterase, CheB.
J.Mol.Biol., 250, 1995
|
|
2CHF
 
 | | STRUCTURE OF THE MG2+-BOUND FORM OF CHEY AND THE MECHANISM OF PHOSPHORYL TRANSFER IN BACTERIAL CHEMOTAXIS | | Descriptor: | CHEY | | Authors: | Stock, A, Martinez-Hackert, E, Rasmussen, B, West, A, Stock, J, Ringe, D, Petsko, G. | | Deposit date: | 1994-01-17 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis.
Biochemistry, 32, 1993
|
|
2CHE
 
 | | STRUCTURE OF THE MG2+-BOUND FORM OF CHEY AND MECHANISM OF PHOSPHORYL TRANSFER IN BACTERIAL CHEMOTAXIS | | Descriptor: | CHEY, MAGNESIUM ION | | Authors: | Stock, A, Martinez-Hackert, E, Rasmussen, B, West, A, Stock, J, Ringe, D, Petsko, G. | | Deposit date: | 1994-01-17 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis.
Biochemistry, 32, 1993
|
|
3C5V
 
 | | PP2A-specific methylesterase apo form (PME) | | Descriptor: | Protein phosphatase methylesterase 1 | | Authors: | Xing, Y, Li, Z, Chen, Y, Stock, J, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural mechanism of demethylation and inactivation of protein phosphatase 2A.
Cell(Cambridge,Mass.), 133, 2008
|
|
3C5W
 
 | | Complex between PP2A-specific methylesterase PME-1 and PP2A core enzyme | | Descriptor: | PP2A A subunit, PP2A C subunit, PP2A-specific methylesterase PME-1 | | Authors: | Xing, Y, Li, Z, Chen, Y, Stock, J, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism of demethylation and inactivation of protein phosphatase 2A.
Cell(Cambridge,Mass.), 133, 2008
|
|
5KX5
 
 | | Crystal structure of tubulin-stathmin-TTL-Compound 11 complex | | Descriptor: | (2~{S},4~{R})-4-[[2-[(1~{R},3~{R})-1-acetyloxy-3-[[(2~{S},3~{S})-2-[[(2~{R})-1,2-dimethylpyrrolidin-2-yl]carbonylamino]-3-methyl-pentanoyl]-methyl-amino]-4-methyl-pentyl]-1,3-thiazol-4-yl]carbonylamino]-5-(4-aminophenyl)-2-methyl-pentanoic acid, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Parris, K. | | Deposit date: | 2016-07-20 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, Synthesis, and Cytotoxic Evaluation of Novel Tubulysin Analogues as ADC Payloads.
ACS Med Chem Lett, 7, 2016
|
|
4H8S
 
 | | Crystal structure of human APPL2BARPH domain | | Descriptor: | DCC-interacting protein 13-beta | | Authors: | Martin, J.L, King, G.J. | | Deposit date: | 2012-09-23 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Membrane Curvature Protein Exhibits Interdomain Flexibility and Binds a Small GTPase.
J.Biol.Chem., 287, 2012
|
|
4ZPE
 
 | |
4ZPF
 
 | |
4ZPG
 
 | |
8DZV
 
 | |
3ZJ6
 
 | | Crystal of Raucaffricine Glucosidase in complex with inhibitor | | Descriptor: | (1R,2S,3S,4R,5R)-4-(cyclohexylmethylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, SULFATE ION | | Authors: | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
2VAQ
 
 | | STRUCTURE OF STRICTOSIDINE SYNTHASE IN COMPLEX WITH INHIBITOR | | Descriptor: | (2S,3R,4S)-methyl 4-(2-(2-(1H-indol-3-yl)ethylamino)ethyl)-2-((2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yloxy)-3-vinyl-3,4-dihydro-2H-pyran-5-carboxylate, STRICTOSIDINE SYNTHASE | | Authors: | Maresh, J, Giddings, L.A, Friedrich, A, Loris, E.A, Panjikar, S, Trout, B.L, Stoeckigt, J, Peters, B, O'Connor, S.E. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Strictosidine synthase: mechanism of a Pictet-Spengler catalyzing enzyme.
J. Am. Chem. Soc., 130, 2008
|
|
6Q4S
 
 | | Crystal structure of a-eudesmol synthase | | Descriptor: | CHLORIDE ION, Pentalenene synthase | | Authors: | Correia Cordeiro, R.S, Hakansson, M, Logan, D.T, Kourist, R. | | Deposit date: | 2018-12-06 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of three novel sesquiterpene synthases from Streptomyces chartreusis NRRL 3882 and crystal structure of an alpha-eudesmol synthase.
J.Biotechnol., 297, 2019
|
|
2V91
 
 | | STRUCTURE OF STRICTOSIDINE SYNTHASE IN COMPLEX WITH STRICTOSIDINE | | Descriptor: | METHYL (2S,3R,4S)-3-ETHYL-2-(BETA-D-GLUCOPYRANOSYLOXY)-4-[(1S)-2,3,4,9-TETRAHYDRO-1H-BETA-CARBOLIN-1-YLMETHYL]-3,4-DIHYDRO-2H-PYRAN-5-CARBOXYLATE, STRICTOSIDINE SYNTHASE | | Authors: | Loris, E.A, Panjikar, S, Ruppert, M, Barleben, L, Unger, M, Stoeckigt, J. | | Deposit date: | 2007-08-16 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structure Based Engineering of Strictosidine Synthase: Auxiliary for Alkaloid Libraries
Chem.Biol., 14, 2007
|
|
2BGH
 
 | | Crystal structure of Vinorine Synthase | | Descriptor: | VINORINE SYNTHASE | | Authors: | Ma, X, Koepke, J, Panjikar, S, Fritzsch, G, Stoeckigt, J. | | Deposit date: | 2004-12-22 | | Release date: | 2005-01-24 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Vinorine Synthase, the First Representative of the Bahd Superfamily.
J.Biol.Chem., 280, 2005
|
|
3GZJ
 
 | | Crystal Structure of Polyneuridine Aldehyde Esterase Complexed with 16-epi-Vellosimine | | Descriptor: | 16-epi-Vellosimine, Polyneuridine-aldehyde esterase | | Authors: | Yang, L, Hill, M, Wang, M, Panjikar, S, Stoeckigt, J. | | Deposit date: | 2009-04-07 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis and enzymatic mechanism of the biosynthesis of C9- from C10-monoterpenoid indole alkaloids
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2WFL
 
 | | Crystal structure of polyneuridine aldehyde esterase | | Descriptor: | POLYNEURIDINE-ALDEHYDE ESTERASE, SULFATE ION | | Authors: | Yang, L, Hill, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2009-04-08 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis and Enzymatic Mechanism of the Biosynthesis of C9- from C10-Monoterpenoid Indole Alkaloids.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
