3J4J
 
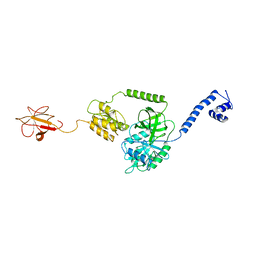 | | Model of full-length T. thermophilus Translation Initiation Factor 2 refined against its cryo-EM density from a 30S Initiation Complex map | | Descriptor: | Translation initiation factor IF-2 | | Authors: | Simonetti, A, Marzi, S, Billas, I.M.L, Tsai, A, Fabbretti, A, Myasnikov, A, Roblin, P, Vaiana, A.C, Hazemann, I, Eiler, D, Steitz, T.A, Puglisi, J.D, Gualerzi, C.O, Klaholz, B.P. | | Deposit date: | 2013-08-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5K1H
 
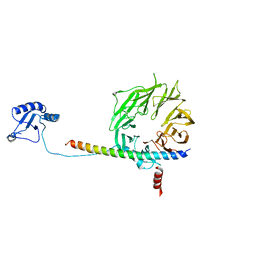 | | eIF3b relocated to the intersubunit face to interact with eIF1 and below the eIF2 ternary-complex. from the structure of a partial yeast 48S preinitiation complex in closed conformation. | | Descriptor: | Eukaryotic translation initiation factor 3 subunit B, eIF3a C-terminal tail | | Authors: | Simonetti, A, Brito Querido, J, Myasnikov, A.G, Mancera-Martinez, E, Renaud, A, Kuhn, L, Hashem, Y. | | Deposit date: | 2016-05-18 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | eIF3 Peripheral Subunits Rearrangement after mRNA Binding and Start-Codon Recognition.
Mol.Cell, 63, 2016
|
|
5K0Y
 
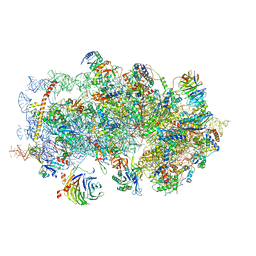 | | m48S late-stage initiation complex, purified from rabbit reticulocytes lysates, displaying eIF2 ternary complex and eIF3 i and g subunits relocated to the intersubunit face | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S12, 40S ribosomal protein S21, ... | | Authors: | Simonetti, A, Brito Querido, J, Myasnikov, A.G, Mancera-Martinez, E, Renaud, A, Kuhn, L, Hashem, Y. | | Deposit date: | 2016-05-17 | | Release date: | 2016-07-13 | | Last modified: | 2018-04-18 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | eIF3 Peripheral Subunits Rearrangement after mRNA Binding and Start-Codon Recognition.
Mol.Cell, 63, 2016
|
|
4B43
 
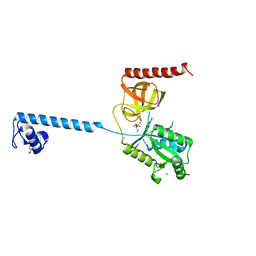 | | Bacterial translation initiation factor IF2 (1-363), apo form, double mutant K86L H130A | | Descriptor: | GLYCEROL, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Simonetti, A, Urzhumtsev, A, Klaholz, B.P. | | Deposit date: | 2012-07-27 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Molecular Basis of GTP Hydrolysis by Bacterial Translation Initiation Factor If2
To be Published
|
|
4B44
 
 | |
4B47
 
 | | Bacterial translation initiation factor IF2 (1-363), complex with GDP at pH6.5 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, TRANSLATION INITIATION FACTOR IF-2 | | Authors: | Simonetti, A, Urzhumtsev, A, Klaholz, B.P. | | Deposit date: | 2012-07-27 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of the Protein Core of Translation Initiation Factor 2 in Apo, GTP-Bound and Gdp-Bound Forms
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4B3X
 
 | | Bacterial translation initiation factor IF2 (1-363), apo form | | Descriptor: | GLYCEROL, TRANSLATION INITIATION FACTOR IF-2 | | Authors: | Simonetti, A, Urzhumtsev, A, Klaholz, B.P. | | Deposit date: | 2012-07-26 | | Release date: | 2013-05-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Protein Core of Translation Initiation Factor 2 in Apo, GTP-Bound and Gdp-Bound Forms
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4B48
 
 | | Bacterial translation initiation factor IF2 (1-363), complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, TRANSLATION INITIATION FACTOR IF-2 | | Authors: | Simonetti, A, Urzhumtsev, A, Klaholz, B.P. | | Deposit date: | 2012-07-27 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Protein Core of Translation Initiation Factor 2 in Apo, GTP-Bound and Gdp-Bound Forms
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6YAM
 
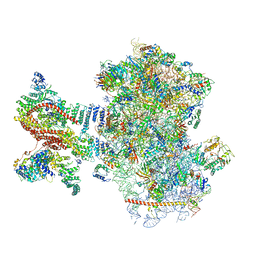 | | Mammalian 48S late-stage translation initiation complex (LS48S+eIF3 IC) with beta-globin mRNA | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein eS1, 40S ribosomal protein eS10, ... | | Authors: | Bochler, A, Simonetti, A, Guca, E, Hashem, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-04-08 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Insights into the Mammalian Late-Stage Initiation Complexes.
Cell Rep, 31, 2020
|
|
6YAN
 
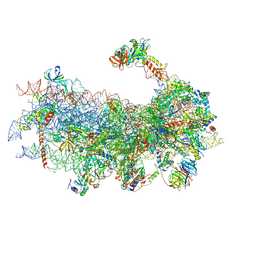 | | Mammalian 48S late-stage translation initiation complex with histone 4 mRNA | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S12, 40S ribosomal protein S21, ... | | Authors: | Bochler, A, Simonetti, A, Guca, E, Hashem, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-04-08 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural Insights into the Mammalian Late-Stage Initiation Complexes.
Cell Rep, 31, 2020
|
|
6YAL
 
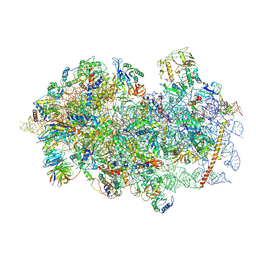 | | Mammalian 48S late-stage initiation complex with beta-globin mRNA | | Descriptor: | 18S ribosomal RNA, 40S Ribosomal protein uS3, 40S Ribosomal protein uS7, ... | | Authors: | Bochler, A, Simonetti, A, Guca, E, Hashem, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-04-08 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Insights into the Mammalian Late-Stage Initiation Complexes.
Cell Rep, 31, 2020
|
|
8CRB
 
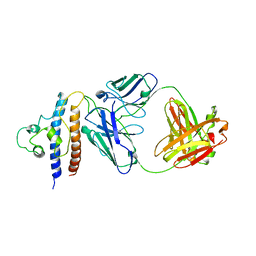 | | Cryo-EM structure of PcrV/Fab(11-E5) | | Descriptor: | Heavy chain, Light chain, Maltose/maltodextrin-binding periplasmic protein,Type III secretion protein PcrV | | Authors: | Yuan, B, Simonis, A, Marlovits, T.C. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Discovery of highly neutralizing human antibodies targeting Pseudomonas aeruginosa.
Cell, 186, 2023
|
|
8CR9
 
 | | Cryo-EM structure of PcrV/Fab(30-B8) | | Descriptor: | Heavy chain, Maltose/maltodextrin-binding periplasmic protein,Type III secretion protein PcrV, light chain | | Authors: | Yuan, B, Simonis, A, Marlovits, T.C. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Discovery of highly neutralizing human antibodies targeting Pseudomonas aeruginosa.
Cell, 186, 2023
|
|
3S64
 
 | | Saposin-like protein Ac-SLP-1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRIC ACID, Saposin-like protein 1 | | Authors: | Willis, C, Wang, C.K, Osman, A, Simon, A, Mulvenna, J, Pickering, D, Riboldi-Tunicliffe, A, Jones, M.K, Loukas, A, Hofmann, A. | | Deposit date: | 2011-05-25 | | Release date: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Insights into the membrane interactions of the saposin-like proteins Na-SLP-1 and Ac-SLP-1 from human and dog hookworm.
Plos One, 6, 2011
|
|
3S63
 
 | | Saposin-like protein Na-SLP-1 | | Descriptor: | Saposin-like protein | | Authors: | Willis, C, Wang, C.K, Osman, A, Simon, A, Mulvenna, J, Pickering, D, Riboldi-Tunicliffe, A, Jones, M.K, Loukas, A, Hofmann, A. | | Deposit date: | 2011-05-24 | | Release date: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the membrane interactions of the saposin-like proteins Na-SLP-1 and Ac-SLP-1 from human and dog hookworm.
Plos One, 6, 2011
|
|
6HNK
 
 | | The ligand-free, open structure of CD0873, a substrate binding protein with adhesive properties from Clostridium difficile. | | Descriptor: | ABC-type transport system, sugar-family extracellular solute-binding protein | | Authors: | Bradshaw, W.J, Kovacs-Simon, A, Harmer, N.J, Michell, S.L, Acharya, K.R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular features of lipoprotein CD0873: A potential vaccine against the human pathogenClostridioides difficile.
J.Biol.Chem., 294, 2019
|
|
6HNJ
 
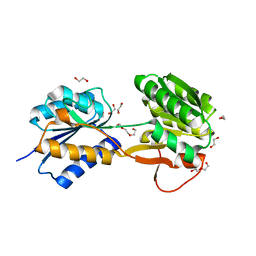 | | The ligand-bound, open structure of CD0873, a substrate binding protein with adhesive properties from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, ABC-type transport system, sugar-family extracellular solute-binding protein, ... | | Authors: | Bradshaw, W.J, Kovacs-Simon, A, Harmer, N.J, Michell, S.L, Acharya, K.R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular features of lipoprotein CD0873: A potential vaccine against the human pathogenClostridioides difficile.
J.Biol.Chem., 294, 2019
|
|
6HNI
 
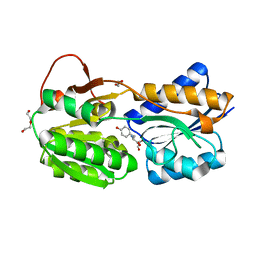 | | The ligand-bound, closed structure of CD0873, a substrate binding protein with adhesive properties from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, ABC-type transport system, sugar-family extracellular solute-binding protein, ... | | Authors: | Bradshaw, W.J, Kovacs-Simon, A, Harmer, N.J, Michell, S.L, Acharya, K.R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Molecular features of lipoprotein CD0873: A potential vaccine against the human pathogenClostridioides difficile.
J.Biol.Chem., 294, 2019
|
|
2MM3
 
 | | Solution NMR structure of the ternary complex of human ileal bile acid-binding protein with glycocholate and glycochenodeoxycholate | | Descriptor: | GLYCOCHENODEOXYCHOLIC ACID, GLYCOCHOLIC ACID, Gastrotropin | | Authors: | Horvath, G, Egyed, O, Bencsura, A, Simon, A, Tochtrop, G.P, DeKoster, G.T, Covey, D.F, Cistola, D.P, Toke, O. | | Deposit date: | 2014-03-07 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of ligand binding in the ternary complex of human ileal bile acid binding protein with glycocholate and glycochenodeoxycholate obtained from solution NMR
FEBS J., 283, 2016
|
|
3ZPK
 
 | | Atomic-resolution structure of a quadruplet cross-beta amyloid fibril. | | Descriptor: | TRANSTHYRETIN | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S.A, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY, SOLID-STATE NMR | | Cite: | Atomic Structure and Hierarchical Assembly of a Cross-Beta Amyloid Fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5E27
 
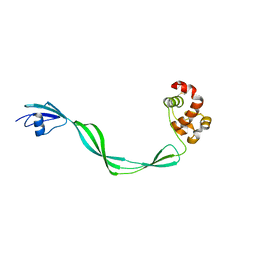 | | The structure of Resuscitation Promoting Factor B from M. tuberculosis reveals unexpected ubiquitin-like domains | | Descriptor: | Resuscitation-promoting factor RpfB | | Authors: | Ruggiero, A, Squeglia, F, Romano, M, Vitagliano, L, De Simone, A, Berisio, R. | | Deposit date: | 2015-09-30 | | Release date: | 2015-11-18 | | Last modified: | 2015-12-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of Resuscitation promoting factor B from M. tuberculosis reveals unexpected ubiquitin-like domains.
Biochim.Biophys.Acta, 1860, 2015
|
|
8RI9
 
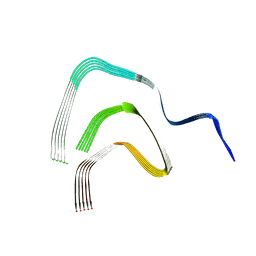 | | Late alpha-Synuclein fibril structure from liquid-liquid phase separations. | | Descriptor: | Alpha-synuclein | | Authors: | De Simone, A, Barritt, J.D, Chen, S, Cascella, R, Cecchi, C, Bigi, A, Jarvis, J.A, Chiti, F, Dobson, C.M, Fusco, G. | | Deposit date: | 2023-12-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-Toxicity Relationship in Intermediate Fibrils from alpha-Synuclein Condensates.
J.Am.Chem.Soc., 146, 2024
|
|
4EMN
 
 | | Crystal structure of RpfB catalytic domain in complex with benzamidine | | Descriptor: | BENZAMIDINE, Probable resuscitation-promoting factor rpfB, SULFATE ION | | Authors: | Ruggiero, A, Marchant, J, Squeglia, F, Makarov, V, De Simone, A, Berisio, R. | | Deposit date: | 2012-04-12 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Molecular determinants of inactivation of the resuscitation promoting factor B from Mycobacterium tuberculosis.
J.Biomol.Struct.Dyn., 31, 2013
|
|
6GS6
 
 | | Cyclophilin A single mutant D66A in complex with an inhibitor. | | Descriptor: | DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase A, ethyl N-[(4-aminobenzyl)carbamoyl]glycinate | | Authors: | Georgiou, C, De Simone, A, Juarez-Jimenez, J, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-06-13 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Dynamic design: manipulation of millisecond timescale motions on the energy landscape of cyclophilin A
Chem Sci, 2020
|
|
2LPF
 
 | | R state structure of monomeric phospholamban (C36A, C41F, C46A) | | Descriptor: | Cardiac phospholamban | | Authors: | De Simone, A, Montalvao, R.W, Gustavsson, M, Shi, L, Veglia, G, Vendruscolo, M. | | Deposit date: | 2012-02-11 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of the Excited States of Phospholamban and Shifts in Their Populations upon Phosphorylation.
Biochemistry, 52, 2013
|
|
