5IVC
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N3 (4'-[(2-phenylethyl)carbamoyl][2,2'-bipyridine]-4-carboxylic acid) | | Descriptor: | 1,2-ETHANEDIOL, 4'-[(2-phenylethyl)carbamoyl][2,2'-bipyridine]-4-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IVF
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N10 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol | | Descriptor: | 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol, Lysine-specific demethylase 5A, MANGANESE (II) ION | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IWF
 
 | |
5IVV
 
 | |
5IVJ
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N11 [3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid] | | Descriptor: | 3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-21 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.569 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IVY
 
 | |
5IVE
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N8 ( 5-methyl-7-oxo-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile) | | Descriptor: | 5-methyl-7-oxo-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, Lysine-specific demethylase 5A, MANGANESE (II) ION | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IW0
 
 | |
4E5D
 
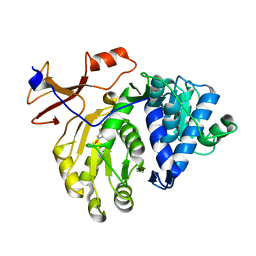 | | 2.2A resolution structure of a firefly luciferase-benzothiazole inhibitor complex | | Descriptor: | 2-(2-fluorophenyl)-6-methoxy-1,3-benzothiazole, Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Throne, N, Shen, M, Auld, D.S, Inglese, J. | | Deposit date: | 2012-03-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter.
Chem.Biol., 19, 2012
|
|
2PU4
 
 | |
2PU2
 
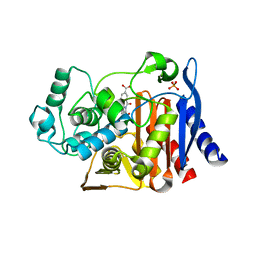 | | AmpC beta-lactamase with bound Phthalamide inhibitor | | Descriptor: | 2-[(1R)-1-CARBOXY-2-(4-HYDROXYPHENYL)ETHYL]-1,3-DIOXOISOINDOLINE-5-CARBOXYLIC ACID, Beta-lactamase, PHOSPHATE ION | | Authors: | Babaoglu, K, Shoichet, B.K. | | Deposit date: | 2007-05-08 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comprehensive mechanistic analysis of hits from high-throughput and docking screens against beta-lactamase.
J.Med.Chem., 51, 2008
|
|
2R9X
 
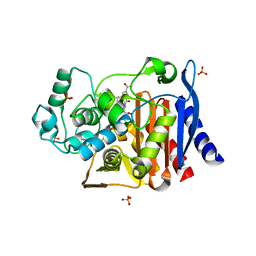 | | AmpC beta-lactamase with bound Phthalamide inhibitor | | Descriptor: | 2-[(1R)-2-carboxy-1-(naphthalen-1-ylmethyl)ethyl]-1,3-dioxo-2,3-dihydro-1H-isoindole-5-carboxylic acid, Beta-lactamase, DIMETHYL SULFOXIDE, ... | | Authors: | Babaoglu, K, Shoichet, B.K. | | Deposit date: | 2007-09-13 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comprehensive mechanistic analysis of hits from high-throughput and docking screens against beta-lactamase.
J.Med.Chem., 51, 2008
|
|
2R9W
 
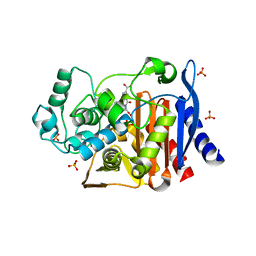 | | AmpC beta-lactamase with bound Phthalamide inhibitor | | Descriptor: | 2-[(1R)-1-carboxy-2-naphthalen-1-ylethyl]-1,3-dioxo-2,3-dihydro-1H-isoindole-5-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Babaoglu, K, Shoichet, B.K. | | Deposit date: | 2007-09-13 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comprehensive mechanistic analysis of hits from high-throughput and docking screens against beta-lactamase.
J.Med.Chem., 51, 2008
|
|
4KW6
 
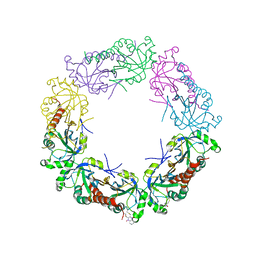 | |
3I06
 
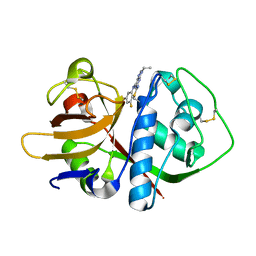 | | Crystal structure of cruzain covalently bound to a purine nitrile | | Descriptor: | 6-[(3,5-difluorophenyl)amino]-9-ethyl-9H-purine-2-carbonitrile, Cruzipain | | Authors: | Ferreira, R.S, Shoichet, B.K, McKerrow, J.H. | | Deposit date: | 2009-06-24 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Identification and optimization of inhibitors of trypanosomal cysteine proteases: cruzain, rhodesain, and TbCatB.
J.Med.Chem., 53, 2010
|
|
3KKU
 
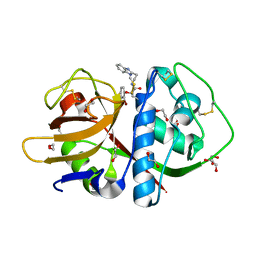 | | Cruzain in complex with a non-covalent ligand | | Descriptor: | 1,2-ETHANEDIOL, Cruzipain, N-[2-(1H-benzimidazol-2-yl)ethyl]-2-(2-bromophenoxy)acetamide, ... | | Authors: | Ferreira, R.S, Eidam, O, Shoichet, B.K. | | Deposit date: | 2009-11-06 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Complementarity between a docking and a high-throughput screen in discovering new cruzain inhibitors.
J.Med.Chem., 53, 2010
|
|
4PE7
 
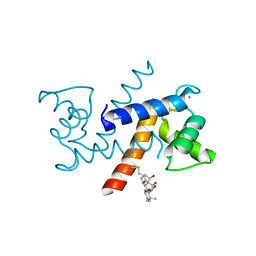 | | Crystal Structure of Calcium-loaded S100B bound to SC1982 | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Pierce, A.D, Wilder, P.T, Neau, D, Toth, E.A, Weber, D.J. | | Deposit date: | 2014-04-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Covalent Small Molecule Inhibitors of Ca(2+)-Bound S100B.
Biochemistry, 53, 2014
|
|
4PE1
 
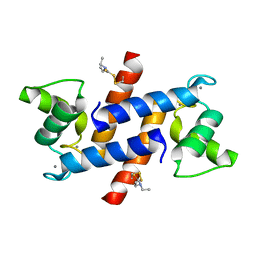 | | Crystal Structure of Calcium-loaded S100B bound to SC124 | | Descriptor: | CALCIUM ION, DIETHYLCARBAMODITHIOIC ACID, Protein S100-B | | Authors: | Cavalier, M.C, Pierce, A.D, Wilder, P.T, Neau, D, Toth, E.A, Weber, D.J. | | Deposit date: | 2014-04-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.576 Å) | | Cite: | Covalent Small Molecule Inhibitors of Ca(2+)-Bound S100B.
Biochemistry, 53, 2014
|
|
4PE0
 
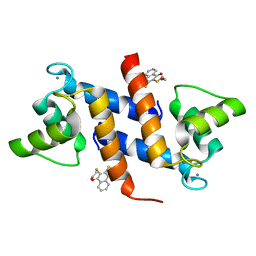 | | Crystal Structure of Calcium-loaded S100B bound to SBi4434 | | Descriptor: | 2-[(2-hydroxyethyl)sulfanyl]naphthalene-1,4-dione, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Pierce, P.D, Wilder, P.T, Neau, D, Toth, E.A, Weber, D.J. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Covalent Small Molecule Inhibitors of Ca(2+)-Bound S100B.
Biochemistry, 53, 2014
|
|
4PDZ
 
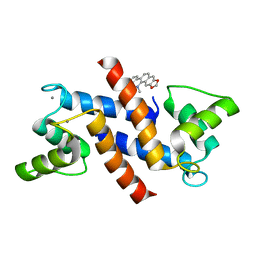 | | Crystal Structure of Calcium-loaded S100B bound to SBi4172 | | Descriptor: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Pierce, A.D, Wilder, P.T, Neau, D, Toth, E.A, Weber, D.J. | | Deposit date: | 2014-04-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Covalent Small Molecule Inhibitors of Ca(2+)-Bound S100B.
Biochemistry, 53, 2014
|
|
4PE4
 
 | | Crystal Structure of Calcium-loaded S100B bound to SC1475 | | Descriptor: | 2,3-dimethoxy-5-[(1S)-1-phenylpropyl]benzene-1,4-diol, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Pierce, A.D, Wilder, P.T, Neau, D, Toth, E.A, Weber, D.J. | | Deposit date: | 2014-04-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.178 Å) | | Cite: | Covalent Small Molecule Inhibitors of Ca(2+)-Bound S100B.
Biochemistry, 53, 2014
|
|
6FP4
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 1,8-Naphthyridine-2-carboxylic acid | | Descriptor: | (2R)-2-hydroxy-3-[4-(2-hydroxyethyl)piperazin-1-yl]propane-1-sulfonic acid, 1,8-naphthyridine-2-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Silvestri, I, Fata, F, MIele, A.E, Boumis, G, Williams, D.L, Angelucci, F. | | Deposit date: | 2018-02-09 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Fragment-Based Discovery of a Regulatory Site in Thioredoxin Glutathione Reductase Acting as "Doorstop" for NADPH Entry.
ACS Chem. Biol., 13, 2018
|
|
6FMZ
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 1,4-Bis(2-hydroxyethyl)piperazine | | Descriptor: | 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanol, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin glutathione reductase | | Authors: | Silvestri, I, Fata, F, MIele, A.E, Boumis, G, Williams, D.L, Angelucci, F. | | Deposit date: | 2018-02-02 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Discovery of a Regulatory Site in Thioredoxin Glutathione Reductase Acting as "Doorstop" for NADPH Entry.
ACS Chem. Biol., 13, 2018
|
|
6FTC
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Silvestri, I, Fata, F, MIele, A.E, Boumis, G, Williams, D.L, Angelucci, F. | | Deposit date: | 2018-02-21 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Discovery of a Regulatory Site in Thioredoxin Glutathione Reductase Acting as "Doorstop" for NADPH Entry.
ACS Chem. Biol., 13, 2018
|
|
6FMU
 
 | | Thioredoxin glutathione reductase from Schistosome mansoni in complex with 2-[4-(4-amino-butyl)-piperazin-1-yl]-ethanol | | Descriptor: | 2-[4-(4-azanylbutyl)piperazin-1-yl]ethanol, FLAVIN-ADENINE DINUCLEOTIDE, TRIETHYLENE GLYCOL, ... | | Authors: | Silvestri, I, Fata, F, MIele, A.E, Boumis, G, Williams, D.L, Angelucci, F. | | Deposit date: | 2018-02-02 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Discovery of a Regulatory Site in Thioredoxin Glutathione Reductase Acting as "Doorstop" for NADPH Entry.
ACS Chem. Biol., 13, 2018
|
|
