6NKL
 
 | | 2.2 A resolution structure of VapBC-1 from nontypeable Haemophilus influenzae | | Descriptor: | Antitoxin VapB1, Ribonuclease VapC | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Molinaro, A.L, Daines, D.A. | | Deposit date: | 2019-01-07 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of VapBC-1 from Nontypeable Haemophilus influenzae and the Effect of PIN Domain Mutations on Survival during Infection.
J.Bacteriol., 201, 2019
|
|
5GRD
 
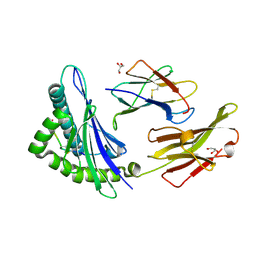 | | Crystal structure of 10-mer peptide from EBV in complex with HLA-A1101. | | Descriptor: | Beta-2-microglobulin, Epstein Barr Virus, Latent membrane protein 2 epitope, ... | | Authors: | Tadwal, V.S, Xiao, Z, Ren, E.C. | | Deposit date: | 2016-08-10 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dual non-contiguous peptide occupancy of HLA class I evoke antiviral human CD8 T cell response and form neo-epitopes with self-antigens
Sci Rep, 7, 2017
|
|
5GSD
 
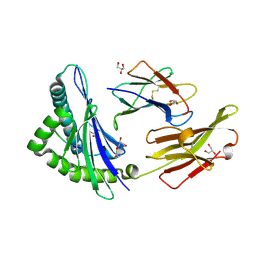 | | Crystal structure of LMP2 peptide from EBV in complex with HLA-A*11:01 | | Descriptor: | Beta-2-microglobulin, Epstein Barr Virus, Latent membrane protein-2 epitope, ... | | Authors: | Tadwal, V.S, Xiao, Z, Ren, E.C. | | Deposit date: | 2016-08-15 | | Release date: | 2017-08-09 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dual non-contiguous peptide occupancy of HLA class I evoke antiviral human CD8 T cell response and form neo-epitopes with self-antigens
Sci Rep, 7, 2017
|
|
5GRG
 
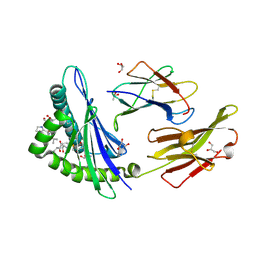 | | Crystal structure of dual peptide from EBV in complex with HLA-A*11:01 | | Descriptor: | Beta-2-microglobulin, Epstein Barr Virus, Latent Membrane Protein 2 epitope, ... | | Authors: | Tadwal, V.S, Xiao, Z, Ren, E.C. | | Deposit date: | 2016-08-11 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Dual non-contiguous peptide occupancy of HLA class I evoke antiviral human CD8 T cell response and form neo-epitopes with self-antigens
Sci Rep, 7, 2017
|
|
6UX9
 
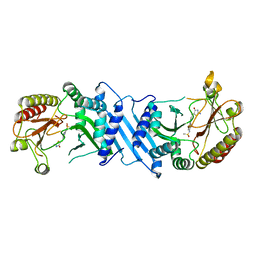 | | Crystal Structure Analysis of PIP4K2A | | Descriptor: | N-[4-(5-{(Z)-[(2E)-2-imino-4-oxo-1,3-thiazolidin-5-ylidene]methyl}pyridin-3-yl)phenyl]methanesulfonamide, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-11-07 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery and Structure-Activity Relationship Study of ( Z )-5-Methylenethiazolidin-4-one Derivatives as Potent and Selective Pan-phosphatidylinositol 5-Phosphate 4-Kinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
3ME3
 
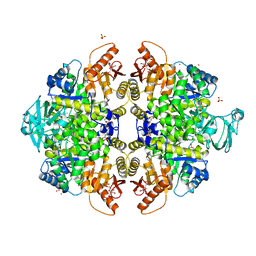 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-{[4-(2,3-dihydro-1,4-benzodioxin-6-ylsulfonyl)-1,4-diazepan-1-yl]sulfonyl}aniline, Pyruvate kinase isozymes M1/M2, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis.
Nat.Chem.Biol., 8, 2012
|
|
5K72
 
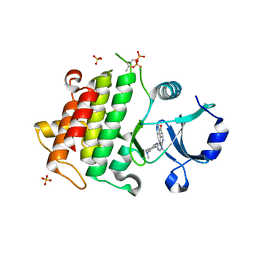 | | IRAK4 in complex with Compound 21 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, ~{N}4,~{N}4-dimethyl-~{N}1-[5-(oxan-4-yl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]cyclohexane-1,4-diamine | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-05-25 | | Release date: | 2017-12-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
5K7G
 
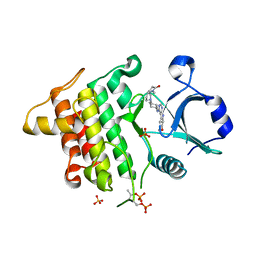 | | IRAK4 in complex with AZ3862 | | Descriptor: | (3~{a}~{S},7~{a}~{R})-1-methyl-5-[4-[[5-(oxan-4-yl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]amino]cyclohexyl]-3,3~{a},4,6,7,7~{a}-hexahydropyrrolo[3,2-c]pyridin-2-one, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-05-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
3U2Z
 
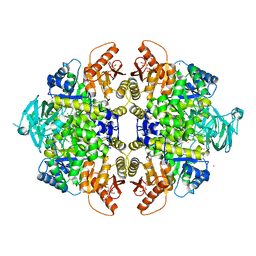 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-(3-aminobenzyl)-4-methyl-2-methylsulfinyl-4,6-dihydro-5H-thieno[2',3':4,5]pyrrolo[2,3-d]pyridazin-5-one, Pyruvate kinase isozymes M1/M2, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-04 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis.
Nat.Chem.Biol., 8, 2012
|
|
8IR4
 
 | | Crystal structure of the SLF1 BRCT domain in complex with a Rad18 peptide containing pS442 | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Xiang, S, Huang, W, Qiu, F. | | Deposit date: | 2023-03-17 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural insights into Rad18 targeting by the SLF1 BRCT domains.
J.Biol.Chem., 299, 2023
|
|
8IR2
 
 | | Crystal structure of the SLF1 BRCT domain in complex with a Rad18 peptide containing pS442 and pS444 | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Xiang, S, Huang, W, Qiu, F. | | Deposit date: | 2023-03-17 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into Rad18 targeting by the SLF1 BRCT domains.
J.Biol.Chem., 299, 2023
|
|
3I06
 
 | | Crystal structure of cruzain covalently bound to a purine nitrile | | Descriptor: | 6-[(3,5-difluorophenyl)amino]-9-ethyl-9H-purine-2-carbonitrile, Cruzipain | | Authors: | Ferreira, R.S, Shoichet, B.K, McKerrow, J.H. | | Deposit date: | 2009-06-24 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Identification and optimization of inhibitors of trypanosomal cysteine proteases: cruzain, rhodesain, and TbCatB.
J.Med.Chem., 53, 2010
|
|
7FDG
 
 | | SARS-COV-2 Spike RBDMACSp6 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDI
 
 | | SARS-COV-2 Spike RBDMACSp36 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDH
 
 | | SARS-COV-2 Spike RBDMACSp25 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDK
 
 | | SARS-COV-2 Spike RBDMACSp36 binding to mACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
4QMG
 
 | | The Structure of MTDH-SND1 Complex Reveals Novel Cancer-Promoting Interactions | | Descriptor: | CESIUM ION, GLYCEROL, Protein LYRIC, ... | | Authors: | Guo, F, Stanevich, V, Wan, L, Satyshur, K, Kang, Y, Xing, Y. | | Deposit date: | 2014-06-16 | | Release date: | 2014-10-08 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural Insights into the Tumor-Promoting Function of the MTDH-SND1 Complex.
Cell Rep, 8, 2014
|
|
6JBR
 
 | | Tps1/UDP/T6P complex | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, Trehalose-6-phosphate synthase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, S, Zhao, Y, Wang, D, Liu, J. | | Deposit date: | 2019-01-26 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
6JAK
 
 | | OtsA apo structure | | Descriptor: | Trehalose-6-phosphate synthase | | Authors: | Wang, S, Zhao, Y, Wang, D, Liu, J. | | Deposit date: | 2019-01-24 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
6JBI
 
 | | Structure of Tps1 apo structure | | Descriptor: | Trehalose-6-phosphate synthase | | Authors: | Wang, S, Zhao, Y, Yi, L, Wang, D, Liu, J. | | Deposit date: | 2019-01-25 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
6JBW
 
 | | Structure of Tps1/UDP complex | | Descriptor: | Trehalose-6-phosphate synthase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, S, Zhao, Y, Wang, D, Liu, J. | | Deposit date: | 2019-01-26 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
