2VQZ
 
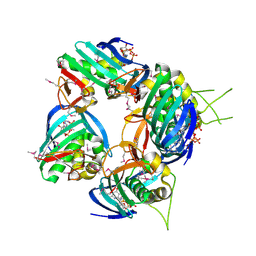 | | Structure of the cap-binding domain of influenza virus polymerase subunit PB2 with bound m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, POLYMERASE BASIC PROTEIN 2 | | Authors: | Guilligay, D, Tarendeau, F, Resa-Infante, P, Coloma, R, Crepin, T, Sehr, P, Lewis, J, Ruigrok, R.W.H, Ortin, J, Hart, D.J, Cusack, S. | | Deposit date: | 2008-03-21 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structural Basis for CAP Binding by Influenza Virus Polymerase Subunit Pb2.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2XFM
 
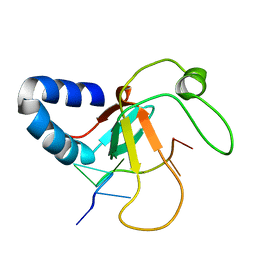 | | Complex structure of the MIWI Paz domain bound to methylated single stranded RNA | | Descriptor: | 5'-R(*AP*CP*CP*GP*AP*CP*UP*(OMU)P)-3', PIWI-LIKE PROTEIN 1 | | Authors: | Simon, B, Kirkpatrick, J.P, Eckhardt, S, Sehr, P, Andrade-Navarro, M.A, Pillai, R.S, Carlomagno, T. | | Deposit date: | 2010-05-27 | | Release date: | 2011-01-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recognition of 2'-O-Methylated 3'-End of Pirna by the Paz Domain of a Piwi Protein.
Structure, 19, 2011
|
|
4CB7
 
 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound cap analogue (compound 8e) | | Descriptor: | 5-[2-(2-azanyl-7-methyl-6-oxidanylidene-1H-purin-9-ium-9-yl)ethanoyl]-2-oxidanyl-benzoic acid, GLYCEROL, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
4CB5
 
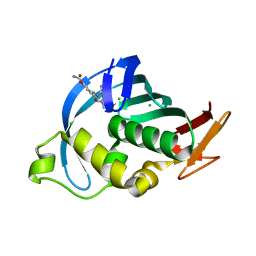 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound cap analogue (compound 8f) | | Descriptor: | 9-N-(3-CARBOXY-4-HYDROXYPHENYL)KETOMETHYL-7-N-METHYLGUANINE, CHLORIDE ION, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
4CB6
 
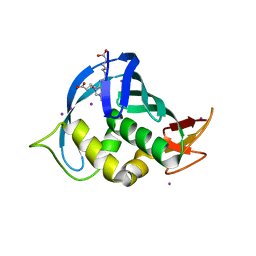 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound cap analogue (compound 11) | | Descriptor: | 2,9-N,N-DI(4-CARBOXYBUTYL)-7-N-METHYLGUANINE, IODIDE ION, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
4CB4
 
 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
7U59
 
 | |
6FHH
 
 | |
6FHI
 
 | |
6VNR
 
 | | Crystal Structure of Danio rerio Histone Deacetylase 6 Catalytic Domain 2 (CD2) Complexed with Bishydroxamic Acid Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Hdac6 protein, N-hydroxy-1-{[4-(hydroxycarbamoyl)phenyl]methyl}-1H-indole-6-carboxamide, ... | | Authors: | Osko, J.D, Porter, N.J, Christianson, D.W. | | Deposit date: | 2020-01-29 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94301319 Å) | | Cite: | Design and Synthesis of Dihydroxamic Acids as HDAC6/8/10 Inhibitors.
Chemmedchem, 15, 2020
|
|
6VNQ
 
 | |
7SMD
 
 | | p107 pocket domain complexed with EID1 peptide | | Descriptor: | EP300-interacting inhibitor of differentiation 1, Retinoblastoma-like protein 1, SULFATE ION | | Authors: | Putta, S, Fernandez, S.M, Tripathi, S.M, Muller, G.A, Rubin, S.M. | | Deposit date: | 2021-10-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for tunable affinity and specificity of LxCxE-dependent protein interactions with the retinoblastoma protein family.
Structure, 30, 2022
|
|
7SME
 
 | | p107 pocket domain complexed with HDAC1 peptide | | Descriptor: | Histone deacetylase 1, Retinoblastoma-like protein 1, SULFATE ION | | Authors: | Putta, S, Fernandez, S.M, Tripathi, S.M, Muller, G.A, Rubin, S.M. | | Deposit date: | 2021-10-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for tunable affinity and specificity of LxCxE-dependent protein interactions with the retinoblastoma protein family.
Structure, 30, 2022
|
|
7SMC
 
 | | p107 pocket domain complexed with ARID4A peptide | | Descriptor: | AT-rich interactive domain-containing protein 4A, Retinoblastoma-like protein 1, SULFATE ION | | Authors: | Putta, S, Fernandez, S.M, Tripathi, S.M, Muller, G.A, Rubin, S.M. | | Deposit date: | 2021-10-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for tunable affinity and specificity of LxCxE-dependent protein interactions with the retinoblastoma protein family.
Structure, 30, 2022
|
|
7SMF
 
 | | p107 pocket domain complexed with mutated HDAC1-3X peptide | | Descriptor: | Histone deacetylase 1, Retinoblastoma-like protein 1, SULFATE ION | | Authors: | Putta, S, Fernandez, S.M, Tripathi, S.M, Muller, G.A, Rubin, S.M. | | Deposit date: | 2021-10-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for tunable affinity and specificity of LxCxE-dependent protein interactions with the retinoblastoma protein family.
Structure, 30, 2022
|
|
7BJS
 
 | | Crystal structure of Khc/atypical Tm1 complex | | Descriptor: | Kinesin heavy chain, SD21996p | | Authors: | Dimitrova-Paternoga, L, Jagtap, P.K.A, Ephrussi, A, Hennig, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Molecular basis of mRNA transport by a kinesin-1-atypical tropomyosin complex.
Genes Dev., 35, 2021
|
|
7BJN
 
 | | Crystal structure of atypical Tm1 (Tm1-I/C), residues 270-334 | | Descriptor: | SD21996p | | Authors: | Dimitrova-Paternoga, L, Jagtap, P.K.A, Ephrussi, A, Hennig, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of mRNA transport by a kinesin-1-atypical tropomyosin complex.
Genes Dev., 35, 2021
|
|
7BJG
 
 | | Crystal structure of atypical Tm1 (Tm1-I/C), residues 262-363 | | Descriptor: | SD21996p | | Authors: | Dimitrova-Paternoga, L, Jagtap, P.K.A, Ephrussi, A, Hennig, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular basis of mRNA transport by a kinesin-1-atypical tropomyosin complex.
Genes Dev., 35, 2021
|
|
3DS2
 
 | | HIV-1 capsid C-terminal domain mutant (Y169A) | | Descriptor: | HIV-1 CAPSID PROTEIN | | Authors: | Vaney, M.-C, Igonet, S, Rey, F.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
3DS5
 
 | | HIV-1 capsid C-terminal domain mutant (N183A) | | Descriptor: | HIV-1 CAPSID PROTEIN | | Authors: | Igonet, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
3DTJ
 
 | | HIV-1 capsid C-terminal domain mutant (E187A) | | Descriptor: | HIV-1 capsid protein | | Authors: | Igonet, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2008-07-15 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
3DS3
 
 | |
3DS4
 
 | |
3DS0
 
 | |
3DPH
 
 | | HIV-1 capsid C-terminal domain mutant (L211S) | | Descriptor: | HIV-1 CAPSID PROTEIN | | Authors: | Igonet, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2008-07-08 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
