1ASY
 
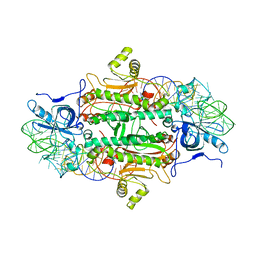 | | CLASS II AMINOACYL TRANSFER RNA SYNTHETASES: CRYSTAL STRUCTURE OF YEAST ASPARTYL-TRNA SYNTHETASE COMPLEXED WITH TRNA ASP | | Descriptor: | ASPARTYL-tRNA SYNTHETASE, T-RNA (75-MER) | | Authors: | Ruff, M, Cavarelli, J, Rees, B, Krishnaswamy, S, Thierry, J.C, Moras, D. | | Deposit date: | 1995-01-19 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp).
Science, 252, 1991
|
|
7BB9
 
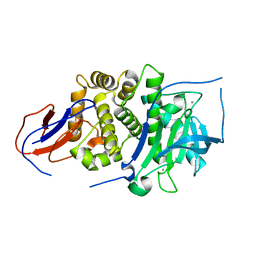 | |
7BB8
 
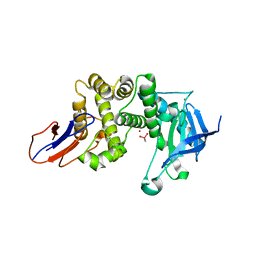 | | Crystal structure of Lugdulysin, a Staphylococcus lugdunensis M30 zinc metallopeptidase | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Ruff, M, Prevost, G, Prola, K, Levy, N. | | Deposit date: | 2020-12-17 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Crystal structure of Staphylococcus lugdunensis protease, Lugdulysin
To Be Published
|
|
1QKT
 
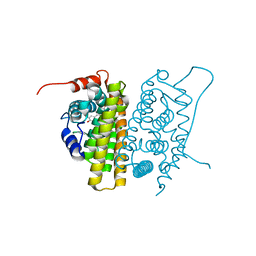 | | MUTANT ESTROGEN NUCLEAR RECEPTOR LIGAND BINDING DOMAIN COMPLEXED WITH ESTRADIOL | | Descriptor: | ESTRADIOL, ESTRADIOL RECEPTOR | | Authors: | Ruff, M, Gangloff, M, Eiler, S, Duclaud, S, Wurtz, J.M, Moras, D. | | Deposit date: | 1999-08-05 | | Release date: | 2000-08-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Mutant Heralpha Ligand- Binding Domain Reveals Key Structural Features for the Mechanism of Partial Agonism
J.Biol.Chem., 276, 2001
|
|
1QKU
 
 | | WILD TYPE ESTROGEN NUCLEAR RECEPTOR LIGAND BINDING DOMAIN COMPLEXED WITH ESTRADIOL | | Descriptor: | ESTRADIOL, ESTRADIOL RECEPTOR | | Authors: | Ruff, M, Gangloff, M, Eiler, S, Duclaud, S, Wurtz, J.M, Moras, D. | | Deposit date: | 1999-08-05 | | Release date: | 2000-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a mutant hERalpha ligand-binding domain reveals key structural features for the mechanism of partial agonism.
J. Biol. Chem., 276, 2001
|
|
6G9P
 
 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-11 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
6T6I
 
 | |
6T6E
 
 | |
6T6J
 
 | |
6G9S
 
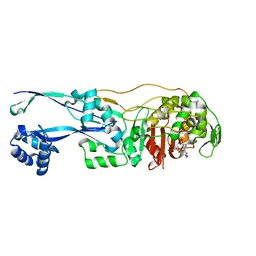 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | (3~{R},6~{S})-6-(aminomethyl)-4-(1,3-oxazol-5-yl)-3-(sulfooxyamino)-3,6-dihydro-2~{H}-pyridine-1-carboxylic acid, Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-11 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
6G9F
 
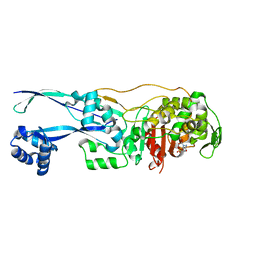 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
4LH5
 
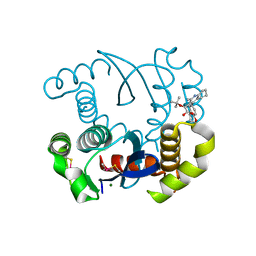 | | Dual inhibition of HIV-1 replication by Integrase-LEDGF allosteric inhibitors is predominant at post-integration stage during virus production rather than at integration | | Descriptor: | (2S)-tert-butoxy[4-(3,4-dihydro-2H-chromen-6-yl)-2-methylquinolin-3-yl]ethanoic acid, Integrase, MAGNESIUM ION | | Authors: | Ruff, M, Levy, N, Eiler, S. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage.
Retrovirology, 10, 2013
|
|
4LH4
 
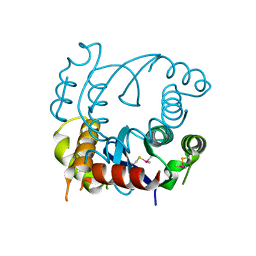 | |
5OI5
 
 | |
5OI8
 
 | |
5OI3
 
 | |
5OIA
 
 | |
5OI2
 
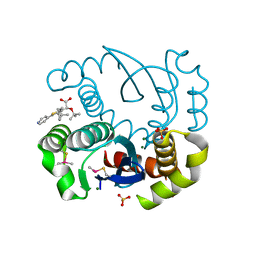 | |
8BV2
 
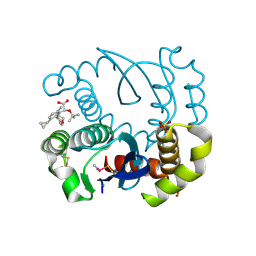 | |
4DM6
 
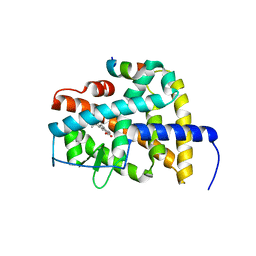 | | Crystal structure of RARb LBD homodimer in complex with TTNPB | | Descriptor: | 4-[(1E)-2-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)PROP-1-ENYL]BENZOIC ACID, Nuclear receptor coactivator 1, Retinoic acid receptor beta | | Authors: | Osz, J, Brelivet, Y, Peluso-Iltis, C, Cura, V, Eiler, S, Ruff, M, Bourguet, W, Rochel, N, Moras, D. | | Deposit date: | 2012-02-07 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3B75
 
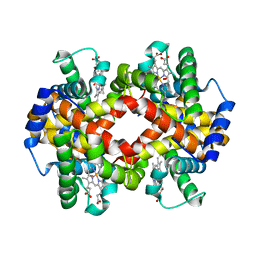 | | Crystal Structure of Glycated Human Haemoglobin | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Saraswathi, N.T, Syakhovich, V.E, Bokut, S.B, Moras, D, Ruff, M. | | Deposit date: | 2007-10-30 | | Release date: | 2008-10-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The effect of hemoglobin glycosylation on diabete linked oxidative stress
To be Published
|
|
4DMA
 
 | | Crystal structure of ERa LBD in complex with RU100132 | | Descriptor: | 2'-bromo-6'-(furan-3-yl)-4'-(hydroxymethyl)biphenyl-4-ol, Estrogen receptor, Nuclear receptor coactivator 1 | | Authors: | Osz, J, Brelivet, Y, Peluso-Iltis, C, Cura, V, Eiler, S, Ruff, M, Bourguet, W, Rochel, N, Moras, D. | | Deposit date: | 2012-02-07 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LBD
 
 | | LIGAND-BINDING DOMAIN OF THE HUMAN RETINOIC ACID RECEPTOR GAMMA BOUND TO ALL-TRANS RETINOIC ACID | | Descriptor: | RETINOIC ACID, RETINOIC ACID RECEPTOR GAMMA | | Authors: | Renaud, J.-P, Rochel, N, Ruff, M, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1997-08-19 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid.
Nature, 378, 1995
|
|
1HV5
 
 | | CRYSTAL STRUCTURE OF THE STROMELYSIN-3 (MMP-11) CATALYTIC DOMAIN COMPLEXED WITH A PHOSPHINIC INHIBITOR | | Descriptor: | 1-BENZYLOXYCARBONYLAMINO-2-PHENYL-ETHYL)-{2-[1-CARBAMOYL-2-(1H-INDOL-3-YL)-ETHYLCARBAMOYL]-5-PHENYL-PENTYL}-PHOSPHINIC ACID, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CALCIUM ION, ... | | Authors: | Gall, A.L, Ruff, M, Kannan, R, Cuniasse, P, Yiotakis, A, Dive, V, Rio, M.C, Basset, P, Moras, D. | | Deposit date: | 2001-01-08 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the stromelysin-3 (MMP-11) catalytic domain complexed with a phosphinic inhibitor mimicking the transition-state.
J.Mol.Biol., 307, 2001
|
|
4DM8
 
 | | Crystal structure of RARb LBD in complex with 9cis retinoic acid | | Descriptor: | Nuclear receptor coactivator 1, RETINOIC ACID, Retinoic acid receptor beta | | Authors: | Osz, J, Br livet, Y, Peluso-Iltis, C, Cura, V, Eiler, S, Ruff, M, Bourguet, W, Rochel, N, Moras, D. | | Deposit date: | 2012-02-07 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
