8TIQ
 
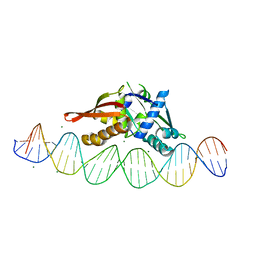 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with blunt ends and 3' terminal phosphates. | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-3', DNA (5'-D(*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Slaughter, C.K, Shrestha, R, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Tuning chemical DNA ligation within DNA crystals and protein-DNA co-crystals
To Be Published
|
|
8TJ0
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 2 sticky base overhangs and 5' terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Slaughter, C.K, Shrestha, R, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Tuning chemical DNA ligation within DNA crystals and co-crystals.
To Be Published
|
|
8TIV
 
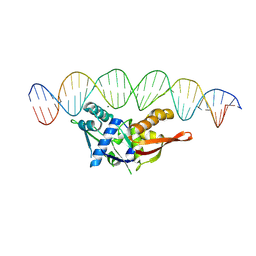 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with blunt ends and 5' terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3')|, DNA (5'-D(A*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Tuning chemical DNA ligation within DNA crystals and co-crystals.
To Be Published
|
|
6MYV
 
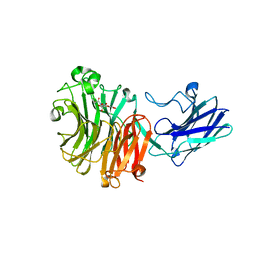 | | Sialidase26 co-crystallized with DANA-Gc | | Descriptor: | 2,6-anhydro-3,5-dideoxy-5-[(hydroxyacetyl)amino]-D-glycero-L-altro-non-2-enonic acid, Sialidase26 | | Authors: | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Huang, J, Siegel, D, Chang, G, Varki, A, Zengler, K. | | Deposit date: | 2018-11-02 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
6OPJ
 
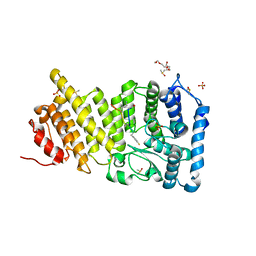 | | Menin in complex with peptide inhibitor 25 | | Descriptor: | DIMETHYL SULFOXIDE, Menin, Peptide inhibitor 25, ... | | Authors: | Linhares, B.M, Fortuna, P, Cierpicki, T, Grembecka, J, Berlicki, L. | | Deposit date: | 2019-04-25 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5006572 Å) | | Cite: | Covalent and noncovalent constraints yield a figure eight-like conformation of a peptide inhibiting the menin-MLL interaction.
Eur.J.Med.Chem., 207, 2020
|
|
6P0J
 
 | | Crystal structure of GDP-bound human RalA | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Ral-A | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PHJ
 
 | |
6PHO
 
 | |
6PHK
 
 | |
6PHP
 
 | |
6PQ7
 
 | |
6PHN
 
 | |
6PHI
 
 | |
6PHM
 
 | |
6P7M
 
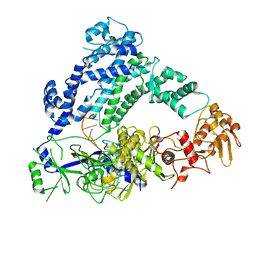 | | Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (1:2 complex) | | Descriptor: | Cas12a, MAGNESIUM ION, anti-CRISPR VA4, ... | | Authors: | Knott, G.J, Liu, J.J, Doudna, J.A. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a.
Elife, 8, 2019
|
|
6PHL
 
 | |
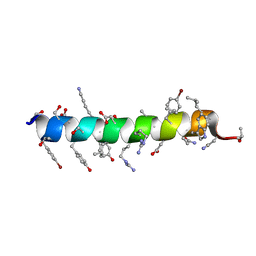
6PHQ
 
 | |
6P7N
 
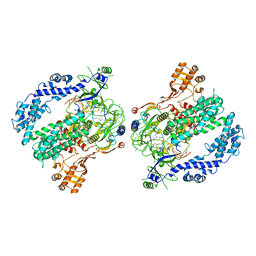 | | Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (2:2 complex) | | Descriptor: | Cas12a, MAGNESIUM ION, anti-CRISPR VA4, ... | | Authors: | Knott, G.J, Liu, J.J, Doudna, J.A. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-21 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a.
Elife, 8, 2019
|
|
6PW8
 
 | |
7PJH
 
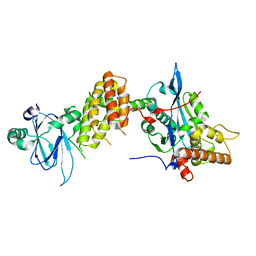 | | Crystal structure of the human spliceosomal maturation factor AAR2 bound to the RNAse H domain of PRPF8 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, Protein AAR2 homolog | | Authors: | Preussner, M, Santos, K, Heroven, A.C, Alles, J, Heyd, F, Wahl, M.C, Weber, G. | | Deposit date: | 2021-08-24 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional investigation of the human snRNP assembly factor AAR2 in complex with the RNase H-like domain of PRPF8.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PQE
 
 | |
7PTH
 
 | |
7PV9
 
 | |
7PQN
 
 | | Catalytic fragment of MASP-2 in complex with ecotin | | Descriptor: | Ecotin, GLYCEROL, Mannan-binding lectin serine protease 2 A chain, ... | | Authors: | Harmat, V, Fodor, K, Heja, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.400015 Å) | | Cite: | Synergy of protease-binding sites within the ecotin homodimer is crucial for inhibition of MASP enzymes and for blocking lectin pathway activation.
J.Biol.Chem., 298, 2022
|
|
7PQO
 
 | | Catalytic fragment of MASP-1 in complex with P1 site mutant ecotin | | Descriptor: | Ecotin, GLYCEROL, Mannan-binding lectin serine protease 1, ... | | Authors: | Harmat, V, Fodor, K, Heja, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Synergy of protease-binding sites within the ecotin homodimer is crucial for inhibition of MASP enzymes and for blocking lectin pathway activation.
J.Biol.Chem., 298, 2022
|
|
