2B9L
 
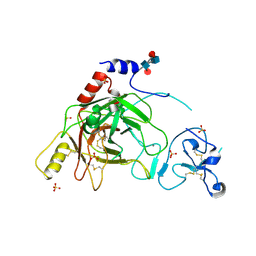 | | Crystal structure of prophenoloxidase activating factor-II from the beetle Holotrichia diomphalia | | Descriptor: | CALCIUM ION, SULFATE ION, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Piao, S, Song, Y.-L, Park, S.Y, Lee, B.L, Oh, B.-H, Ha, N.-C. | | Deposit date: | 2005-10-12 | | Release date: | 2006-01-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a clip-domain serine protease and functional roles of the clip domains.
Embo J., 24, 2005
|
|
7FCI
 
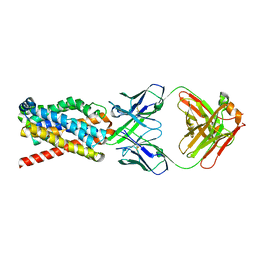 | | human NTCP in complex with YN69083 Fab | | Descriptor: | Fab Heavy chain, Fab Light chain, Sodium/bile acid cotransporter | | Authors: | Park, J.H, Iwamoto, M, Yun, J.H, Uchikubo-Kamo, T, Son, D, Jin, Z, Yoshida, H, Ohki, M, Ishimoto, N, Mizutani, K, Oshima, M, Muramatsu, M, Wakita, T, Shirouzu, M, Liu, K, Uemura, T, Nomura, N, Iwata, S, Watashi, K, Tame, J.R.H, Nishizawa, T, Lee, W, Park, S.Y. | | Deposit date: | 2021-07-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the HBV receptor and bile acid transporter NTCP.
Nature, 606, 2022
|
|
8IC0
 
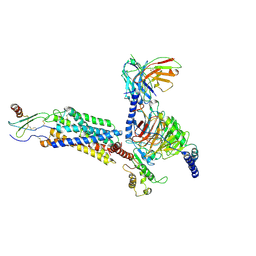 | | Cryo-EM structure of CXCL8 bound C-X-C chemokine receptor 1 in complex with Gi heterotrimer | | Descriptor: | C-X-C chemokine receptor type 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ishimoto, N, Park, J.H, Park, S.Y. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural basis of CXC chemokine receptor 1 ligand binding and activation.
Nat Commun, 14, 2023
|
|
8I7W
 
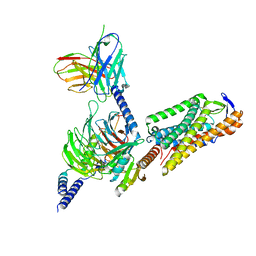 | | Cryo-EM structure of GSK256073 bound human hydroxy-carboxylic acid receptor 2 in complex with Gi heterotrimer | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Park, J.H, Ishimoto, N, Park, S.Y. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis for ligand recognition and signaling of hydroxy-carboxylic acid receptor 2.
Nat Commun, 14, 2023
|
|
8I7V
 
 | | Cryo-EM structure of Acipimox bound human hydroxy-carboxylic acid receptor 2 in complex with Gi heterotrimer | | Descriptor: | 5-methyl-4-oxidanyl-pyrazin-4-ium-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Park, J.H, Ishimoto, N, Park, S.Y. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for ligand recognition and signaling of hydroxy-carboxylic acid receptor 2.
Nat Commun, 14, 2023
|
|
8K5D
 
 | |
8K5B
 
 | |
8K5C
 
 | |
1JFB
 
 | | X-ray structure of nitric oxide reductase (cytochrome P450nor) in the ferric resting state at atomic resolution | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, nitric-oxide reductase cytochrome P450 55A1 | | Authors: | Shimizu, H, Adachi, S, Park, S.Y, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-06-20 | | Release date: | 2001-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray structure of nitric oxide reductase (cytochrome P450nor) at atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1JFC
 
 | | X-ray structure of nitric oxide reductase (cytochrome P450nor) in the ferrous CO state at atomic resolution | | Descriptor: | CARBON MONOXIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shimizu, H, Adachi, S, Park, S.Y, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-06-20 | | Release date: | 2001-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | X-ray structure of nitric oxide reductase (cytochrome P450nor) at atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4YED
 
 | | TcdA (CsdL) | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, tRNA threonylcarbamoyladenosine dehydratase | | Authors: | Kim, S, Park, S.Y. | | Deposit date: | 2015-02-24 | | Release date: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of Escherichia coli TcdA (Also Known As CsdL) Reveals a Novel Topology and Provides Insight into the tRNA Binding Surface Required for N(6)-Threonylcarbamoyladenosine Dehydratase Activity
J.Mol.Biol., 427, 2015
|
|
4YH8
 
 | | Structure of yeast U2AF complex | | Descriptor: | Splicing factor U2AF 23 kDa subunit, Splicing factor U2AF 59 kDa subunit, ZINC ION | | Authors: | Yoshida, H, Park, S.Y, Urano, T, Obayashi, E. | | Deposit date: | 2015-02-27 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel 3' splice site recognition by the two zinc fingers in the U2AF small subunit.
Genes Dev., 29, 2015
|
|
3JTH
 
 | |
2D5Z
 
 | | Crystal structure of T-state human hemoglobin complexed with three L35 molecules | | Descriptor: | 2-[4-({[(3,5-DICHLOROPHENYL)AMINO]CARBONYL}AMINO)PHENOXY]-2-METHYLPROPANOIC ACID, Hemoglobin alpha subunit, Hemoglobin beta subunit, ... | | Authors: | Yokoyama, T, Neya, S, Tsuneshige, A, Yonetani, T, Park, S.Y, Tame, J.R. | | Deposit date: | 2005-11-08 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | R-state haemoglobin with low oxygen affinity: crystal structures of deoxy human and carbonmonoxy horse haemoglobin bound to the effector molecule L35
J.Mol.Biol., 356, 2006
|
|
2D60
 
 | | Crystal structure of deoxy human hemoglobin complexed with two L35 molecules | | Descriptor: | 2-[4-({[(3,5-DICHLOROPHENYL)AMINO]CARBONYL}AMINO)PHENOXY]-2-METHYLPROPANOIC ACID, Hemoglobin alpha subunit, Hemoglobin beta subunit, ... | | Authors: | Yokoyama, T, Neya, S, Tsuneshige, A, Yonetani, T, Park, S.Y, Tame, J.R. | | Deposit date: | 2005-11-08 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | R-state haemoglobin with low oxygen affinity: crystal structures of deoxy human and carbonmonoxy horse haemoglobin bound to the effector molecule L35
J.Mol.Biol., 356, 2006
|
|
2D5X
 
 | | Crystal structure of carbonmonoxy horse hemoglobin complexed with L35 | | Descriptor: | 2-[4-({[(3,5-DICHLOROPHENYL)AMINO]CARBONYL}AMINO)PHENOXY]-2-METHYLPROPANOIC ACID, CARBON MONOXIDE, Hemoglobin alpha subunit, ... | | Authors: | Yokoyama, T, Neya, S, Tsuneshige, A, Yonetani, T, Park, S.Y, Tame, J.R. | | Deposit date: | 2005-11-08 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | R-state haemoglobin with low oxygen affinity: crystal structures of deoxy human and carbonmonoxy horse haemoglobin bound to the effector molecule L35
J.Mol.Biol., 356, 2006
|
|
2EB4
 
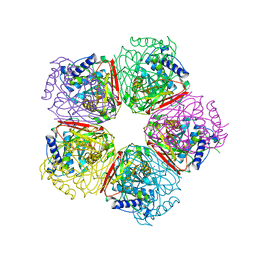 | | Crystal structure of apo-HpcG | | Descriptor: | 2-oxo-hept-3-ene-1,7-dioate hydratase, SODIUM ION, THIOCYANATE ION | | Authors: | Izumi, A, Rea, D, Adachi, T, Unzai, S, Park, S.Y, Roper, D.I, Tame, J.R.H. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Mechanism of HpcG, a Hydratase in the Homoprotocatechuate Degradation Pathway of Escherichia coli
J.Mol.Biol., 370, 2007
|
|
2EB5
 
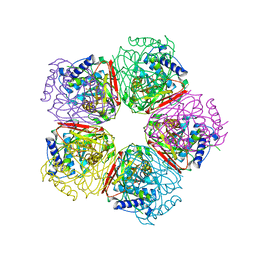 | | Crystal structure of HpcG complexed with oxalate | | Descriptor: | 2-oxo-hept-3-ene-1,7-dioate hydratase, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Izumi, A, Rea, D, Adachi, T, Unzai, S, Park, S.Y, Roper, D.I, Tame, J.R.H. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Mechanism of HpcG, a Hydratase in the Homoprotocatechuate Degradation Pathway of Escherichia coli
J.Mol.Biol., 370, 2007
|
|
2EB6
 
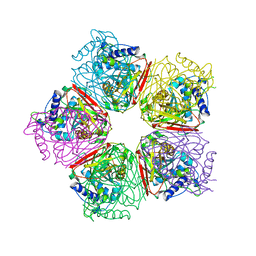 | | Crystal structure of HpcG complexed with Mg ion | | Descriptor: | 2-oxo-hept-3-ene-1,7-dioate hydratase, MAGNESIUM ION | | Authors: | Izumi, A, Rea, D, Adachi, T, Unzai, S, Park, S.Y, Roper, D.I, Tame, J.R.H. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure and Mechanism of HpcG, a Hydratase in the Homoprotocatechuate Degradation Pathway of Escherichia coli
J.Mol.Biol., 370, 2007
|
|
2EXS
 
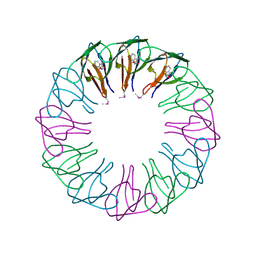 | | TRAP3 (engineered TRAP) | | Descriptor: | TRYPTOPHAN, Transcription attenuation protein mtrB | | Authors: | Heddle, J.G, Yokoyama, T, Yamashita, I, Park, S.Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rounding up: Engineering 12-Membered Rings from the Cyclic 11-Mer TRAP
Structure, 14, 2006
|
|
1L8O
 
 | | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase | | Descriptor: | L-3-phosphoserine phosphatase, PHOSPHATE ION, SERINE | | Authors: | Kim, H.Y, Heo, Y.S, Kim, J.H, Park, M.H, Moon, J, Park, S.Y, Lee, T.G, Jeon, Y.H, Ro, S, Hwang, K.Y. | | Deposit date: | 2002-03-21 | | Release date: | 2003-04-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase
J.Biol.Chem., 277, 2002
|
|
1L8L
 
 | | Molecular basis for the local confomational rearrangement of human phosphoserine phosphatase | | Descriptor: | D-2-AMINO-3-PHOSPHONO-PROPIONIC ACID, L-3-phosphoserine phosphatase | | Authors: | Kim, H.Y, Heo, Y.S, Kim, J.H, Park, M.H, Moon, J, Park, S.Y, Lee, T.G, Jeon, Y.H, Ro, S, Hwang, K.Y. | | Deposit date: | 2002-03-21 | | Release date: | 2003-04-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase.
J.Biol.Chem., 277, 2002
|
|
2F9Z
 
 | | Complex between the chemotaxis deamidase CheD and the chemotaxis phosphatase CheC from Thermotoga maritima | | Descriptor: | PROTEIN (chemotaxis methylation protein), chemotaxis protein CheC | | Authors: | Chao, X, Park, S.Y, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2005-12-06 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation.
Cell(Cambridge,Mass.), 124, 2006
|
|
2EXT
 
 | | TRAP4 (engineered TRAP) | | Descriptor: | TRYPTOPHAN, Transcription attenuation protein mtrB | | Authors: | Heddle, J.G, Yokoyama, T, Yamashita, I, Park, S.Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rounding up: Engineering 12-Membered Rings from the Cyclic 11-Mer TRAP
Structure, 14, 2006
|
|
2IHS
 
 | |
