5WBS
 
 | | Crystal structure of Frizzled-7 CRD with an inhibitor peptide Fz7-21 | | Descriptor: | Frizzled-7,inhibitor peptide Fz7-21 | | Authors: | Nile, A.H, Mukund, S, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-06-29 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
5W96
 
 | | Solution structure of phage derived peptide inhibitor of frizzled 7 receptor | | Descriptor: | Fz7 binding peptide | | Authors: | Nile, A.H, de Sousa e Melo, F, Mukund, S, Piskol, R, Hansen, S, Zhou, L, Zhang, Y, Fu, Y, Gogol, E.B, Komuves, L.G, Modrusan, Z, Angers, S, Franke, Y, Koth, C, Fairbrother, W.J, Wang, W, de Sauvage, F.J, Hannoush, R.N. | | Deposit date: | 2017-06-22 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
5T44
 
 | | Crystal structure of Frizzled 7 CRD | | Descriptor: | Frizzled-7 | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannous, R.H, Wang, W. | | Deposit date: | 2016-08-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9944 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5URV
 
 | | Crystal structure of Frizzled 7 CRD in complex with C24 fatty acid | | Descriptor: | (15E)-TETRACOS-15-ENOIC ACID, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5URZ
 
 | | Crystal structure of Frizzled 5 CRD in complex with BOG | | Descriptor: | Frizzled-5, alpha-L-fucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, octyl beta-D-glucopyranoside | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5URY
 
 | | Crystal structure of Frizzled 5 CRD in complex with PAM | | Descriptor: | Frizzled-5, PALMITOLEIC ACID, alpha-L-fucopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8GCR
 
 | | HPV16 E6-E6AP-p53 complex | | Descriptor: | Cellular tumor antigen p53, Maltose/maltodextrin-binding periplasmic protein,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Bratkowski, M.A, Wang, J.C.K, Hao, Q, Nile, A.H. | | Deposit date: | 2023-03-02 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structure of the p53 degradation complex from HPV16.
Nat Commun, 15, 2024
|
|
7UFG
 
 | | Cryo-EM structure of PAPP-A in complex with IGFBP5 | | Descriptor: | Insulin-like growth factor-binding protein 5, Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-03-22 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
8D8O
 
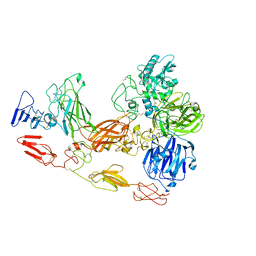 | | Cryo-EM structure of substrate unbound PAPP-A | | Descriptor: | Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-06-08 | | Release date: | 2022-09-28 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
4M8Z
 
 | |
7NAM
 
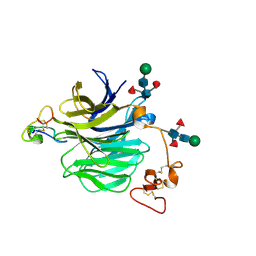 | | LRP6_E1 in complex with Lr-EET-3.5 | | Descriptor: | Low-density lipoprotein receptor-related protein 6, SODIUM ION, Trypsin inhibitor 2, ... | | Authors: | Hansen, S, Hannoush, R.N. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Directed evolution identifies high-affinity cystine-knot peptide agonists and antagonists of Wnt/ beta-catenin signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZGB
 
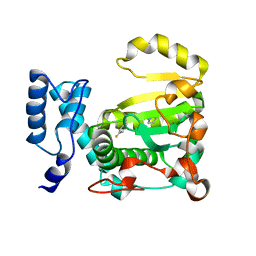 | | Structure of yeast Sec14p with NPPM112 | | Descriptor: | 4-fluoranyl-~{N}-[(4-pyrrolidin-1-ylphenyl)methyl]benzamide, SEC14 cytosolic factor | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
7ZG9
 
 | | Structure of yeast Sec14p with himbacine | | Descriptor: | Himbacine, SEC14 cytosolic factor | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
7ZGA
 
 | | Structure of yeast Sec14p with ergoline | | Descriptor: | SEC14 cytosolic factor, ~{O}9-methyl ~{O}4-[2,2,2-tris(chloranyl)ethyl] (5~{a}~{S},6~{a}~{S},9~{R},10~{a}~{S})-7-methyl-3-nitro-5,5~{a},6,6~{a},8,9,10,10~{a}-octahydroindolo[4,3-fg]quinoline-4,9-dicarboxylate | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
7ZGD
 
 | | Structure of yeast Sec14p with NPPM244 | | Descriptor: | (4-bromanyl-3-nitro-phenyl)-[4-(2-fluorophenyl)piperazin-1-yl]methanone, SEC14 cytosolic factor | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
7ZGC
 
 | | Structure of yeast Sec14p with NPPM481 | | Descriptor: | (4-chloranyl-3-nitro-phenyl)-[4-(2-fluorophenyl)piperazin-1-yl]methanone, PHOSPHATE ION, SEC14 cytosolic factor | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
