1U58
 
 | | Crystal structure of the murine cytomegalovirus MHC-I homolog m144 | | 分子名称: | MHC-I homolog m144, beta-2-microglobulin | | 著者 | Natarajan, K, Hicks, A, Robinson, H, Guan, R, Margulies, D.H. | | 登録日 | 2004-07-27 | | 公開日 | 2005-07-19 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the murine cytomegalovirus MHC-I homolog m144.
J.Mol.Biol., 358, 2006
|
|
5IVX
 
 | | Crystal Structure of B4.2.3 T-Cell Receptor and H2-Dd P18-I10 Complex | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | 著者 | Natarajan, K, Jiang, J, Margulies, D. | | 登録日 | 2016-03-21 | | 公開日 | 2017-03-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | An allosteric site in the T-cell receptor C beta domain plays a critical signalling role.
Nat Commun, 8, 2017
|
|
5IW1
 
 | |
1FM5
 
 | |
3ECB
 
 | | Crystal structure of mouse H-2Dd in complex with peptide P18-I10 derived from human immunodeficiency virus envelope glycoprotein 120 | | 分子名称: | 1,2-ETHANEDIOL, Beta-2 microglobulin, H-2 class I histocompatibility antigen, ... | | 著者 | Natarajan, K, Wang, R, Margulies, D.H. | | 登録日 | 2008-08-29 | | 公開日 | 2009-07-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.698 Å) | | 主引用文献 | Structural basis of the CD8alphabeta/MHC class i interaction: focused recognition orients CD8beta to a T cell proximal position
J.Immunol., 183, 2009
|
|
7MFV
 
 | | Crystal structure of synthetic nanobody (Sb16) | | 分子名称: | 1,2-ETHANEDIOL, Synthetic Nanobody #16 (Sb16) | | 著者 | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | 登録日 | 2021-04-11 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7KGK
 
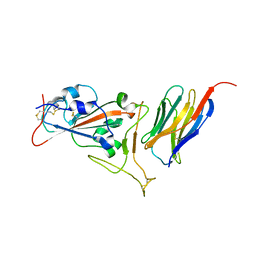 | | Crystal structure of synthetic nanobody (Sb16) complexes with SARS-CoV-2 receptor binding domain | | 分子名称: | Sb16, Sybody-16, Synthetic Nanobody, ... | | 著者 | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | 登録日 | 2020-10-16 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7KLW
 
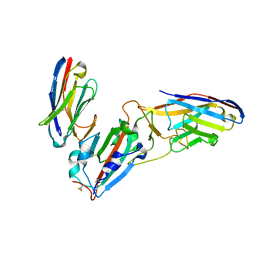 | | Crystal structure of synthetic nanobody (Sb45+Sb68) complexes with SARS-CoV-2 receptor binding domain | | 分子名称: | SB45, Synthetic Nanobody, SB68, ... | | 著者 | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | 登録日 | 2020-11-01 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7KGJ
 
 | | Crystal structure of synthetic nanobody (Sb45) complexes with SARS-CoV-2 receptor binding domain | | 分子名称: | Sb45, Sybody-45, Synthetic Nanobody, ... | | 著者 | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | 登録日 | 2020-10-16 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
1P1Z
 
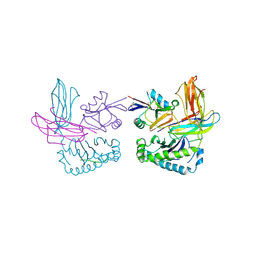 | | X-RAY CRYSTAL STRUCTURE OF THE LECTIN-LIKE NATURAL KILLER CELL RECEPTOR LY-49C BOUND TO ITS MHC CLASS I LIGAND H-2Kb | | 分子名称: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | 著者 | Dimasi, N, Natarajan, K, Rangjin, G, Dam, J, Margulies, D.H, Mariuzza, R.A. | | 登録日 | 2003-04-14 | | 公開日 | 2003-11-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.26 Å) | | 主引用文献 | Variable MHC class I engagement by Ly49 natural killer cell receptors demonstrated by the crystal structure of Ly49C bound to H-2K(b).
Nat.Immunol., 4, 2003
|
|
1JA3
 
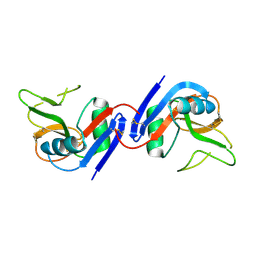 | | Crystal Structure of the Murine NK Cell Inhibitory Receptor Ly-49I | | 分子名称: | MHC class I recognition receptor Ly49I | | 著者 | Dimasi, N, Sawicki, W.M, Reineck, L.A, Li, Y, Natarajan, K, Murgulies, D.H, Mariuzza, A.R. | | 登録日 | 2001-05-29 | | 公開日 | 2002-07-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of the Ly49I natural killer cell receptor reveals variability in dimerization mode within the Ly49 family.
J.Mol.Biol., 320, 2002
|
|
7MFU
 
 | | Crystal structure of synthetic nanobody (Sb14+Sb68) complexes with SARS-CoV-2 receptor binding domain | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Spike protein S1, ... | | 著者 | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | 登録日 | 2021-04-11 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
4G59
 
 | |
2MIZ
 
 | |
2O5N
 
 | | Crystal structure of a Viral Glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MuHV1gpm153, ... | | 著者 | Mans, J, Natarajan, K, Robinson, H, Margulies, D.H. | | 登録日 | 2006-12-06 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Cellular Expression and Crystal Structure of the Murine Cytomegalovirus Major Histocompatibility Complex Class I-like Glycoprotein, m153.
J.Biol.Chem., 282, 2007
|
|
8TQA
 
 | | Crystal structure of Fab.28.14.8 in complex with MHC-I (H2-Db) | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | 著者 | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | 登録日 | 2023-08-06 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQ8
 
 | | Crystal structure of Fab.34.5.8 in complex with MHC-I (H2-Dd) | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab.34.5.8 Heavy chain, ... | | 著者 | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | 登録日 | 2023-08-06 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQ7
 
 | | Crystal structure of Fab.34.2.12 in complex with MHC-I (H2-Dd) | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab 34.2.12 Light Chain, ... | | 著者 | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | 登録日 | 2023-08-06 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQ9
 
 | | Crystal structure of Fab.S19.8 in complex with MHC-I (H2-Dd) | | 分子名称: | Beta-2-microglobulin, Fab.S19.8 Heavy Chain, Fab.S19.8 Light Chain, ... | | 著者 | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | 登録日 | 2023-08-06 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
3E6F
 
 | | MHC CLASS I H-2Dd Heavy chain complexed with Beta-2 Microglobulin and a variant peptide, PA9, from the Human immunodeficiency virus (BaL) envelope glycoprotein 120 | | 分子名称: | BETA-2 MICROGLOBULIN, Envelope glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | 著者 | Wang, R, Natarajan, K, Robinson, H, Margulies, D.H. | | 登録日 | 2008-08-15 | | 公開日 | 2009-08-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Different vaccine vectors delivering the same antigen elicit CD8+ T cell responses with distinct clonotype and epitope specificity
J.Immunol., 183, 2009
|
|
3E6H
 
 | | MHC CLASS I H-2Dd heavy chain complexed with Beta-2 Microglobulin and a variant peptide, PI10, from the human immunodeficiency virus (BaL) envelope glycoprotein 120 | | 分子名称: | Envelope glycoprotein 10-residue peptide, H-2 class I histocompatibility antigen, D-D alpha chain, ... | | 著者 | Wang, R, Natarajan, K, Robinson, H, Margulies, D.H. | | 登録日 | 2008-08-15 | | 公開日 | 2009-08-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Different vaccine vectors delivering the same antigen elicit CD8+ T cell responses with distinct clonotype and epitope specificity
J.Immunol., 183, 2009
|
|
1P4L
 
 | | Crystal structure of NK receptor Ly49C mutant with its MHC class I ligand H-2Kb | | 分子名称: | Beta-2-microglobulin, LY49-C, MHC CLASS I H-2KB HEAVY CHAIN, ... | | 著者 | Dam, J, Guan, R, Natarajan, K, Dimasi, N, Mariuzza, R.A. | | 登録日 | 2003-04-23 | | 公開日 | 2003-11-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Variable MHC class I engagement by Ly49 natural killer cell receptors demonstrated by the crystal structure of Ly49C bound to H-2K(b).
Nat.Immunol., 4, 2003
|
|
3DMM
 
 | | Crystal structure of the CD8 alpha beta/H-2Dd complex | | 分子名称: | Beta-2 microglobulin, H-2 class I histocompatibility antigen, D-D alpha chain, ... | | 著者 | Wang, R, Natarajan, K, Margulies, D.H. | | 登録日 | 2008-07-01 | | 公開日 | 2009-07-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis of the CD8alphabeta/MHC class i interaction: focused recognition orients CD8beta to a T cell proximal position.
J.Immunol., 183, 2009
|
|
5WES
 
 | | Crystal Structure H2-Dd with disulfide-linked 5mer peptide | | 分子名称: | Beta-2-microglobulin, GLYCINE, H-2 class I histocompatibility antigen, ... | | 著者 | Jiang, J.S, Natarajan, K, Boyd, L.F, Margulies, D.H. | | 登録日 | 2017-07-10 | | 公開日 | 2017-10-18 | | 最終更新日 | 2017-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.706 Å) | | 主引用文献 | Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation.
Science, 358, 2017
|
|
5WEU
 
 | | Crystal Structure of H2-Dd with disulfide-linked 10mer peptide | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Envelope glycoprotein gp160, ... | | 著者 | Jiang, J.S, Natarajan, K, Boyd, L.F, Margulies, D.H. | | 登録日 | 2017-07-10 | | 公開日 | 2017-10-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.584 Å) | | 主引用文献 | Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation.
Science, 358, 2017
|
|
