3M45
 
 | | Crystal structure of Ig1 domain of mouse SynCAM 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 2 | | Authors: | Yue, L, Modis, Y. | | Deposit date: | 2010-03-10 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | N-glycosylation at the SynCAM (synaptic cell adhesion molecule) immunoglobulin interface modulates synaptic adhesion.
J.Biol.Chem., 285, 2010
|
|
6T1B
 
 | |
6T0Y
 
 | |
3TS9
 
 | | Crystal Structure of the MDA5 Helicase Insert Domain | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, SULFATE ION | | Authors: | Berke, I.C, Modis, Y. | | Deposit date: | 2011-09-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA.
Embo J., 31, 2012
|
|
6TL1
 
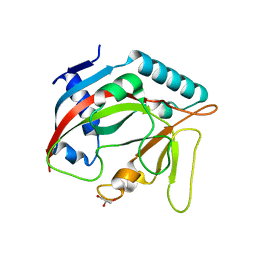 | | Crystal structure of the TASOR pseudo-PARP domain | | Descriptor: | GLYCEROL, Protein TASOR | | Authors: | Douse, C.H, Timms, R.T, Freund, S.M.V, Modis, Y. | | Deposit date: | 2019-11-29 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control.
Nat Commun, 11, 2020
|
|
6QAJ
 
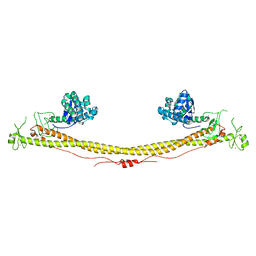 | | Structure of the tripartite motif of KAP1/TRIM28 | | Descriptor: | Endolysin,Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Stoll, G.A, Oda, S, Yu, M, Modis, Y. | | Deposit date: | 2018-12-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structure of KAP1 tripartite motif identifies molecular interfaces required for retroelement silencing.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7NGA
 
 | |
7NIQ
 
 | |
7NIC
 
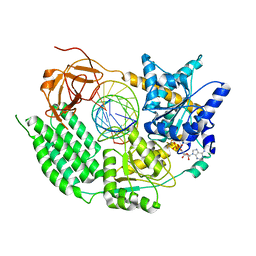 | |
2I69
 
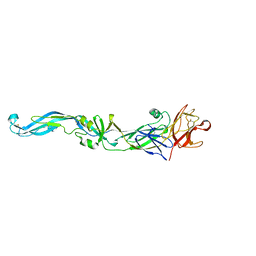 | | Crystal structure of the West Nile virus envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Polyprotein | | Authors: | Kanai, R, Modis, Y. | | Deposit date: | 2006-08-28 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of west nile virus envelope glycoprotein reveals viral surface epitopes.
J.Virol., 80, 2006
|
|
3NEC
 
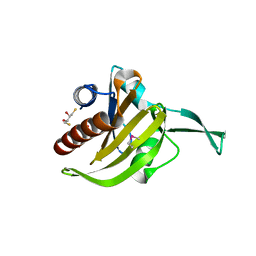 | | Crystal Structure of Toxoplasma gondii Profilin | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Inflammatory profilin, SULFATE ION | | Authors: | Kucera, K, Modis, Y. | | Deposit date: | 2010-06-08 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based analysis of Toxoplasma gondii profilin: a parasite-specific motif is required for recognition by Toll-like receptor 11.
J.Mol.Biol., 403, 2010
|
|
3NSW
 
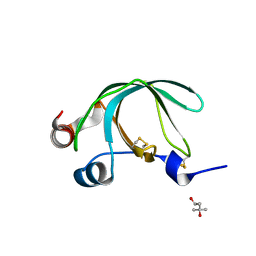 | | Crystal Structure of Ancylostoma ceylanicum Excretory-Secretory Protein 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Excretory-secretory protein 2 | | Authors: | Kucera, K, Modis, Y. | | Deposit date: | 2010-07-02 | | Release date: | 2011-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ancylostoma ceylanicum Excretory-Secretory Protein 2 Adopts a Netrin-Like Fold and Defines a Novel Family of Nematode Proteins.
J.Mol.Biol., 408, 2011
|
|
7BKP
 
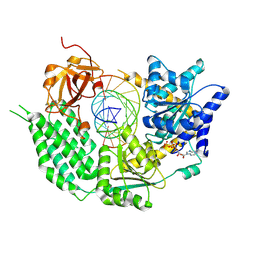 | | CryoEM structure of disease related M854K MDA5-dsRNA filament in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Singh, R, Herrero del Valle, A, Yu, Q, Modis, Y. | | Deposit date: | 2021-01-16 | | Release date: | 2021-11-17 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | MDA5 disease variant M854K prevents ATP-dependent structural discrimination of viral and cellular RNA.
Nat Commun, 12, 2021
|
|
7BKQ
 
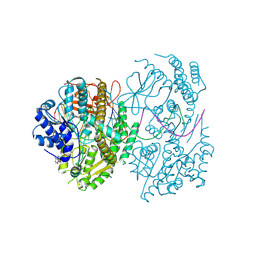 | | CryoEM structure of MDA5-dsRNA filament in complex with ADP with 92-degree helical twist | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Isoform 2 of Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*GP*UP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*GP*A)-3'), ... | | Authors: | Yu, Q, Modis, Y. | | Deposit date: | 2021-01-17 | | Release date: | 2021-11-17 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | MDA5 disease variant M854K prevents ATP-dependent structural discrimination of viral and cellular RNA.
Nat Commun, 12, 2021
|
|
4JNT
 
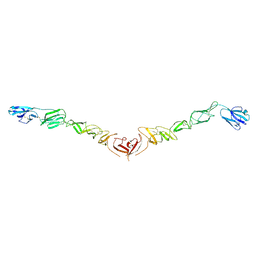 | |
1RH5
 
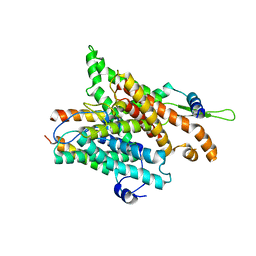 | | The structure of a protein conducting channel | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, SecBeta | | Authors: | van den Berg, B, Clemons Jr, W.M, Collinson, I, Modis, Y, Hartmann, E, Harrison, S.C, Rapoport, T.A. | | Deposit date: | 2003-11-13 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of a protein-conducting channel
Nature, 427, 2004
|
|
1RHZ
 
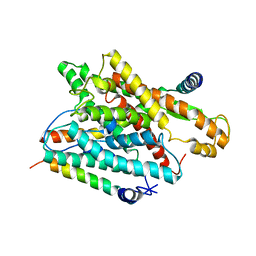 | | The structure of a protein conducting channel | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, SecBeta | | Authors: | van den Berg, B, Clemons Jr, W.M, Collinson, I, Modis, Y, Hartmann, E, Harrison, S.C, Rapoport, T.A. | | Deposit date: | 2003-11-15 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | X-ray structure of a protein-conducting channel.
Nature, 427, 2004
|
|
4DQ7
 
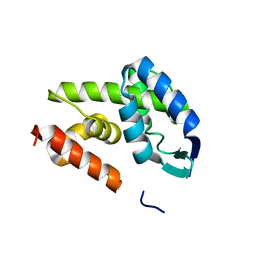 | |
4DQJ
 
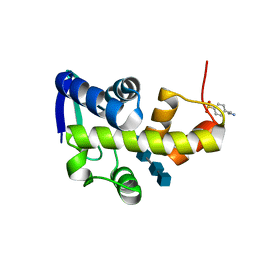 | | Structural Investigation of Bacteriophage Phi6 Lysin (in complex with chitotetraose) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, Membrane protein Phi6 P5 | | Authors: | Dessau, M.A, Modis, Y. | | Deposit date: | 2012-02-16 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Selective pressure causes an RNA virus to trade reproductive fitness for increased structural and thermal stability of a viral enzyme.
PLoS Genet, 8, 2012
|
|
4DQ5
 
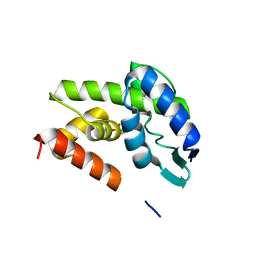 | | Structural Investigation of Bacteriophage Phi6 Lysin (WT) | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, Membrane protein Phi6 P5wt | | Authors: | Dessau, M.A, Modis, Y. | | Deposit date: | 2012-02-15 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Selective pressure causes an RNA virus to trade reproductive fitness for increased structural and thermal stability of a viral enzyme.
PLoS Genet, 8, 2012
|
|
6G1X
 
 | | CryoEM structure of the MDA5-dsRNA filament with 91-degree helical twist | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*GP*UP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*GP*A)-3'), RNA (5'-R(P*UP*CP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*AP*CP*G)-3'), ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-21 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
6G1S
 
 | | CryoEM structure of the MDA5-dsRNA filament with 87-degree helical twist | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*GP*UP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*GP*A)-3'), RNA (5'-R(P*UP*CP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*AP*CP*G)-3'), ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-03-21 | | Release date: | 2018-11-21 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
6G19
 
 | | CryoEM structure of the MDA5-dsRNA filament with 74-degree helical twist | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RNA (5'-R(P*CP*AP*AP*GP*CP*CP*GP*AP*GP*GP*AP*GP*AP*G)-3'), ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-21 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
6GKM
 
 | | CryoEM structure of the MDA5-dsRNA filament in complex with ATP (10 mM) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*AP*AP*GP*CP*CP*GP*AP*GP*GP*AP*GP*AP*G)-3'), ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-05-21 | | Release date: | 2018-11-21 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
6H61
 
 | | CryoEM structure of the MDA5-dsRNA filament with 89 degree twist and without nucleotide | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*GP*UP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*GP*A)-3'), RNA (5'-R(P*UP*CP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*AP*CP*G)-3'), ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-07-25 | | Release date: | 2018-11-21 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
