1PWG
 
 | | Covalent Penicilloyl Acyl Enzyme Complex Of The Streptomyces R61 DD-Peptidase With A Highly Specific Penicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(6S)-6-carboxy-6-(glycylamino)hexanoyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, D-alanyl-D-alanine carboxypeptidase | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.074 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
4C63
 
 | | ULTRA HIGH RESOLUTION DICKERSON-DREW DODECAMER B-DNA WITH 5- METHYLCYSTOSINE MODIFICATION | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*5CMP*GP*CP*GP)-3', MAGNESIUM ION | | Authors: | Mcdonough, M, El-Sagheer, A.H, Brown, T, Schofield, C.J. | | Deposit date: | 2013-09-17 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural insights into how 5-hydroxymethylation influences transcription factor binding.
Chem. Commun. (Camb.), 50, 2014
|
|
5K48
 
 | | VIM-2 Metallo Beta Lactamase in complex with 3-(mercaptomethyl)-[1,1'-biphenyl]-4-carboxylic acid | | Descriptor: | 4-phenyl-2-(sulfanylmethyl)benzoic acid, Beta-lactamase VIM-2, FORMIC ACID, ... | | Authors: | Zollman, D, McDonough, M, Brem, J, Schofield, C. | | Deposit date: | 2016-05-20 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | In Silico Fragment-Based Design Identifies Subfamily B1 Metallo-beta-lactamase Inhibitors.
J. Med. Chem., 61, 2018
|
|
4C64
 
 | | ULTRA HIGH RESOLUTION DICKERSON-DREW DODECAMER B-DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*GP)-3', MAGNESIUM ION | | Authors: | Mcdonough, M, El-Sagheer, A.H, Brown, T, Schofield, C.J. | | Deposit date: | 2013-09-17 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural insights into how 5-hydroxymethylation influences transcription factor binding.
Chem. Commun. (Camb.), 50, 2014
|
|
4C5X
 
 | | Ultra High Resolution Dickerson-Drew dodecamer B-DNA with 5-Hydroxymethyl-cytosine Modification | | Descriptor: | 5'-D(CP*GP*CP*GP*AP*AP*TP*TP*5HCP*GP*CP*GP)-3', MAGNESIUM ION | | Authors: | McDonough, M, El-Sagheer, A.H, Brown, T, Schofield, C.J. | | Deposit date: | 2013-09-16 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insights into how 5-hydroxymethylation influences transcription factor binding.
Chem. Commun. (Camb.), 50, 2014
|
|
5MNU
 
 | | OXA-10 Avibactam complex with bound bromide | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, BROMIDE ION, Beta-lactamase OXA-10, ... | | Authors: | Brem, J, McDonough, M, Clifton, I. | | Deposit date: | 2016-12-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | (13)C-Carbamylation as a mechanistic probe for the inhibition of class D beta-lactamases by avibactam and halide ions.
Org. Biomol. Chem., 15, 2017
|
|
4AI9
 
 | | JMJD2A Complexed with Daminozide | | Descriptor: | CHLORIDE ION, DAMINOZIDE, GLYCEROL, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2012-02-08 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Plant growth regulator daminozide is a selective inhibitor of human KDM2/7 histone demethylases.
J. Med. Chem., 55, 2012
|
|
4BIS
 
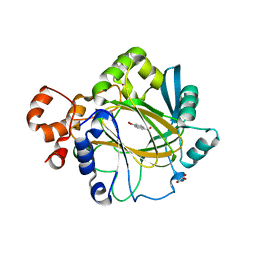 | | JMJD2A COMPLEXED WITH 8-HYDROXYQUINOLINE-4-CARBOXYLIC ACID | | Descriptor: | 8-hydroxyquinoline-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chowdhury, R, Thinnes, C, Schofield, C.J. | | Deposit date: | 2013-04-12 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | 5-Carboxy-8-hydroxyquinoline is a Broad Spectrum 2-Oxoglutarate Oxygenase Inhibitor which Causes Iron Translocation.
Chem Sci, 4, 2013
|
|
4BIO
 
 | |
4B7E
 
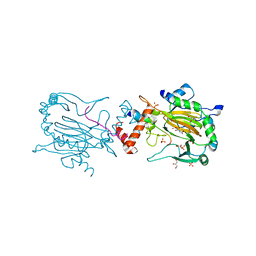 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH CONSENSUS ANKYRIN REPEAT DOMAIN-LEU PEPTIDE (20-MER) | | Descriptor: | CONSENSUS ANKYRIN REPEAT DOMAIN-LEU, GLYCEROL, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, ... | | Authors: | Chowdhury, R, Ge, W, Schofield, C.J. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate selectivity analyses of factor inhibiting hypoxia-inducible factor.
Angew. Chem. Int. Ed. Engl., 52, 2013
|
|
4B7K
 
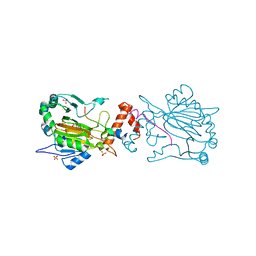 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH CONSENSUS ANKYRIN REPEAT DOMAIN-SER PEPTIDE (20-MER) | | Descriptor: | CONSENSUS ANKYRIN REPEAT DOMAIN-SER, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, N-OXALYLGLYCINE, ... | | Authors: | Chowdhury, R, Ge, W, Schofield, C.J. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Substrate selectivity analyses of factor inhibiting hypoxia-inducible factor.
Angew. Chem. Int. Ed. Engl., 52, 2013
|
|
4BQX
 
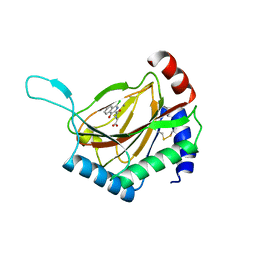 | |
7A5Z
 
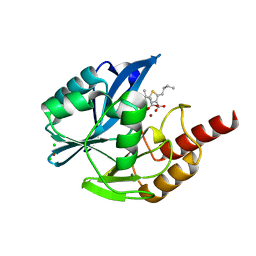 | | Structure of VIM-2 metallo-beta-lactamase with hydrolysed Faropenem imine product | | Descriptor: | (5~{Z})-2-[1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-5-(4-oxidanylbutylidene)-2~{H}-1,3-thiazole-4-carboxylic acid, Beta-lactamase VIM-2, CHLORIDE ION, ... | | Authors: | Lucic, A, Schofield, C.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Faropenem reacts with serine and metallo-beta-lactamases to give multiple products.
Eur.J.Med.Chem., 215, 2021
|
|
7A60
 
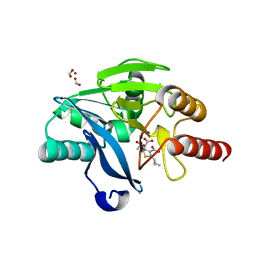 | | Crystal structure of VIM-2 with hydrolyzed faropenem (ring-open form) | | Descriptor: | (5~{Z})-2-[1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-5-(4-oxidanylbutylidene)-2~{H}-1,3-thiazole-4-carboxylic acid, Beta-lactamase VIM-2, FORMIC ACID, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Faropenem reacts with serine and metallo-beta-lactamases to give multiple products.
Eur.J.Med.Chem., 215, 2021
|
|
7A61
 
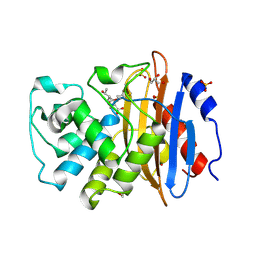 | | Crystal structure of KPC-2 with hydrolyzed faropenem (ring-open form) | | Descriptor: | (2~{R})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-5-butyl-2,3-dihydro-1,3-thiazole-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Faropenem reacts with serine and metallo-beta-lactamases to give multiple products.
Eur.J.Med.Chem., 215, 2021
|
|
7A63
 
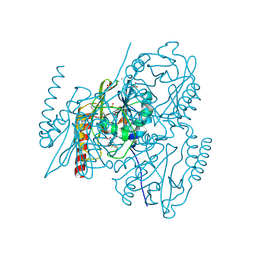 | | Crystal structure of L1 with hydrolyzed faropenem (imine, ring-closed form) | | Descriptor: | (2R,5S)-2-[(1S,2R)-1-carboxy-2-hydroxy-propyl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydrothiazole-4-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57000113 Å) | | Cite: | Faropenem reacts with serine and metallo-beta-lactamases to give multiple products.
Eur.J.Med.Chem., 215, 2021
|
|
7AFZ
 
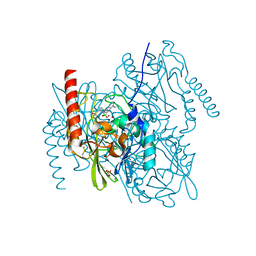 | | L1 metallo-b-lactamase with compound EBL-1306 | | Descriptor: | 3-[3-chloranyl-4-(methylsulfonylmethyl)phenyl]-7-propan-2-yl-1~{H}-indole-2-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J, Brem, J. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Imitation of beta-lactam binding enables broad-spectrum metallo-beta-lactamase inhibitors.
Nat.Chem., 14, 2022
|
|
6H8P
 
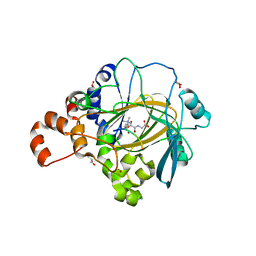 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II), NOG AND Histone H1.4(18-32)K26me3 peptide (15-mer) | | Descriptor: | CHLORIDE ION, GLYCEROL, Histone H1.4, ... | | Authors: | Chowdhury, R, Walport, L.J, Schofield, C.J. | | Deposit date: | 2018-08-03 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Mechanistic and structural studies of KDM-catalysed demethylation of histone 1 isotype 4 at lysine 26.
FEBS Lett., 592, 2018
|
|
4UWD
 
 | |
2XEP
 
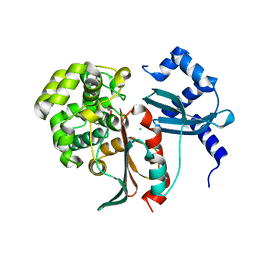 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | CHLORIDE ION, GLYCEROL, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-05-17 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2XFS
 
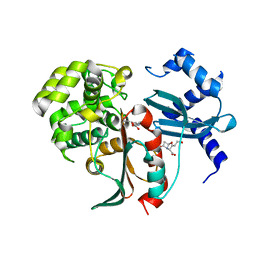 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2XH9
 
 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-06-09 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2XGN
 
 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | GLYCEROL, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-06-07 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2XF3
 
 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-05-20 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2XFT
 
 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | GLYCEROL, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
