7ONS
 
 | | PARP1 catalytic domain in complex with isoquinolone-based inhibitor (compound 16) | | Descriptor: | 7-[[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]methyl]-3-ethyl-1~{H}-quinolin-2-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
7ONR
 
 | | PARP1 catalytic domain in complex with 8-chloroquinazolinone-based inhibitor (compound 9) | | Descriptor: | 8-chloranyl-2-[3-[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]propyl]-3~{H}-quinazolin-4-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
7ONT
 
 | | PARP1 catalytic domain in complex with a selective pyridine carboxamide-based inhibitor (compound 22) | | Descriptor: | 5-[4-[(3-ethyl-2-oxidanylidene-1~{H}-quinolin-7-yl)methyl]piperazin-1-yl]-~{N}-methyl-pyridine-2-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M.J, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
5R4U
 
 | | PanDDA analysis group deposition -- Crystal Structure of HUMAN CLEAVAGE FACTOR IM in complex with EN08775-43 | | Descriptor: | (5S)-5-methyl-5-[(3-phenyl-1,2-oxazol-5-yl)methyl]pyrrolidine-2,4-dione, ACETATE ION, Cleavage and polyadenylation specificity factor subunit 5, ... | | Authors: | Kidd, S.L, Mateu, N, Talon, R, Krojer, T, Aimon, A, Bradley, A.R, Fairhead, M, Diaz-Saez, L, Sore, H.F, Madin, A, Huber, K.V.M, von Delft, F, Spring, D.R. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R4T
 
 | | PanDDA analysis group deposition -- Crystal Structure of HUMAN CLEAVAGE FACTOR IM in complex with SK-430 | | Descriptor: | (5R)-5-{[1-(4-fluorophenyl)-1H-1,2,3-triazol-4-yl]methyl}-5-methylpyrrolidine-2,4-dione, ACETATE ION, Cleavage and polyadenylation specificity factor subunit 5, ... | | Authors: | Kidd, S.L, Mateu, N, Talon, R, Krojer, T, Aimon, A, Bradley, A.R, Fairhead, M, Diaz-Saez, L, Sore, H.F, Madin, A, Huber, K.V.M, von Delft, F, Spring, D.R. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R4R
 
 | | PanDDA analysis group deposition -- Crystal Structure of HUMAN CLEAVAGE FACTOR IM in complex with EN08775-42 | | Descriptor: | (5R)-3,3,5-trimethyl-5-[(1-phenyl-1H-1,2,3-triazol-4-yl)methyl]pyrrolidine-2,4-dione, ACETATE ION, Cleavage and polyadenylation specificity factor subunit 5, ... | | Authors: | Kidd, S.L, Mateu, N, Talon, R, Krojer, T, Aimon, A, Bradley, A.R, Fairhead, M, Diaz-Saez, L, Sore, H.F, Madin, A, Huber, K.V.M, von Delft, F, Spring, D.R. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R4P
 
 | | PanDDA analysis group deposition -- Crystal Structure of HUMAN CLEAVAGE FACTOR IM in complex with NM450-1 | | Descriptor: | (5S)-5-methyl-5-[(1-phenyl-1H-1,2,3-triazol-4-yl)methyl]pyrrolidine-2,4-dione, ACETATE ION, Cleavage and polyadenylation specificity factor subunit 5, ... | | Authors: | Kidd, S.L, Mateu, N, Talon, R, Krojer, T, Aimon, A, Bradley, A.R, Fairhead, M, Diaz-Saez, L, Sore, H.F, Madin, A, Huber, K.V.M, von Delft, F, Spring, D.R. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R4Q
 
 | | PanDDA analysis group deposition -- Crystal Structure of HUMAN CLEAVAGE FACTOR IM in complex with NM466-1 | | Descriptor: | (3S)-3,4-dimethyl-3-propyl-3,4-dihydro-1H-1,4-benzodiazepine-2,5-dione, ACETATE ION, Cleavage and polyadenylation specificity factor subunit 5, ... | | Authors: | Kidd, S.L, Mateu, N, Talon, R, Krojer, T, Aimon, A, Bradley, A.R, Fairhead, M, Diaz-Saez, L, Sore, H.F, Madin, A, Huber, K.V.M, von Delft, F, Spring, D.R. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R4S
 
 | | PanDDA analysis group deposition -- Crystal Structure of HUMAN CLEAVAGE FACTOR IM in complex with EN08775-45 | | Descriptor: | (4S,5R)-4-hydroxy-5-methyl-5-[(1-phenyl-1H-1,2,3-triazol-4-yl)methyl]pyrrolidin-2-one, ACETATE ION, Cleavage and polyadenylation specificity factor subunit 5, ... | | Authors: | Kidd, S.L, Mateu, N, Talon, R, Krojer, T, Aimon, A, Bradley, A.R, Fairhead, M, Diaz-Saez, L, Sore, H.F, Madin, A, Huber, K.V.M, von Delft, F, Spring, D.R. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
8OW2
 
 | | Crystal structure of the p110alpha catalytic subunit from homo sapiens in complex with activator 1938 | | Descriptor: | 1-[7-[[2-[[4-(4-ethylpiperazin-1-yl)phenyl]amino]pyridin-4-yl]amino]-2,3-dihydroindol-1-yl]ethanone, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gong, G.Q, Bellini, D, Vanhaesebroeck, B, Williams, R.L. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
8BFU
 
 | | Crystal structure of the apo p110alpha catalytic subunit from homo sapiens | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gong, G.Q, Bellini, D, Vanhaesebroeck, B, Williams, R.L. | | Deposit date: | 2022-10-26 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
7PG5
 
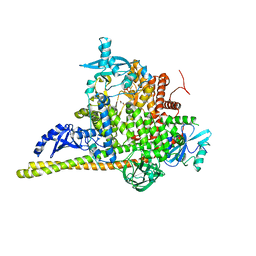 | | Crystal Structure of PI3Kalpha | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gong, G, Pinotsis, N, Williams, R.L, Vanhaesebroeck, B. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.20029068 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
7PG6
 
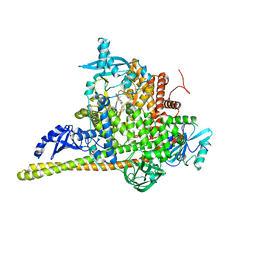 | | Crystal Structure of PI3Kalpha in complex with the inhibitor NVP-BYL719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Gong, G, Pinotsis, N, Williams, R.L, Vanhaesebroeck, B. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49943733 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
8AN8
 
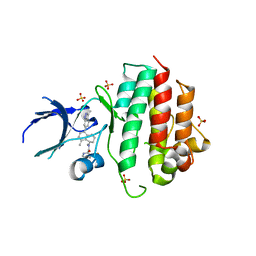 | | Crystal structure of wild-type c-MET bound by compound 7. | | Descriptor: | 3-[bis(fluoranyl)methyl]-~{N}-methyl-~{N}-[(1~{R})-8-methyl-5-(3-methyl-1~{H}-indazol-6-yl)-1,2,3,4-tetrahydronaphthalen-1-yl]pyridine-2-carboxamide, Hepatocyte growth factor receptor, SULFATE ION | | Authors: | Collie, G.W. | | Deposit date: | 2022-08-04 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Discovery of a selective c-MET inhibitor with a novel binding mode.
Bioorg.Med.Chem.Lett., 75, 2022
|
|
8ANS
 
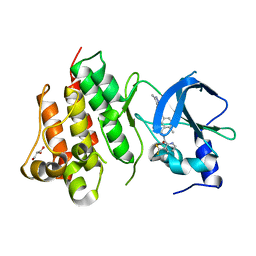 | | Crystal structure of D1228V c-MET bound by compound 1. | | Descriptor: | 3-[bis(fluoranyl)methyl]-~{N}-methyl-~{N}-[(1~{R})-8-methyl-5-(3-methyl-1~{H}-indazol-6-yl)-1,2,3,4-tetrahydronaphthalen-1-yl]pyridine-2-carboxamide, GLYCEROL, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2022-08-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Discovery of a selective c-MET inhibitor with a novel binding mode.
Bioorg.Med.Chem.Lett., 75, 2022
|
|
7R9L
 
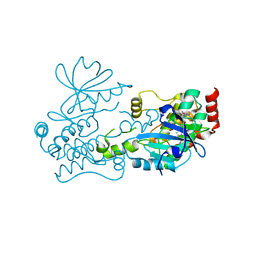 | | Crystal structure of HPK1 in complex with compound 2 | | Descriptor: | 2-amino-N,N-dimethyl-5-(1H-pyrrolo[2,3-b]pyridin-5-yl)benzamide, Hematopoietic progenitor kinase | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
7R9P
 
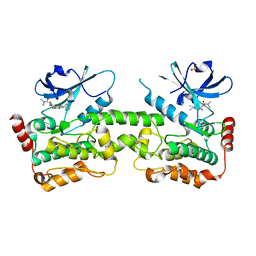 | | Crystal structure of HPK1 in complex with compound 14 | | Descriptor: | 6-amino-2-fluoro-N,N-dimethyl-3-(4'-methylspiro[cyclopropane-1,3'-pyrrolo[2,3-b]pyridin]-5'-yl)benzamide, Hematopoietic progenitor kinase, SULFATE ION | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
7R9T
 
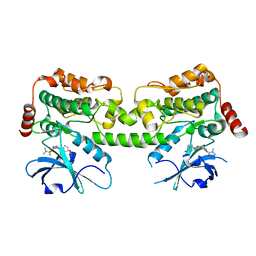 | | Crystal structure of HPK1 in complex with compound 17 | | Descriptor: | 6-amino-3-[(1S,3R)-4'-chloro-3-hydroxy-1',2'-dihydrospiro[cyclopentane-1,3'-pyrrolo[2,3-b]pyridin]-5'-yl]-2-fluoro-N,N-dimethylbenzamide, Hematopoietic progenitor kinase | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
7R9N
 
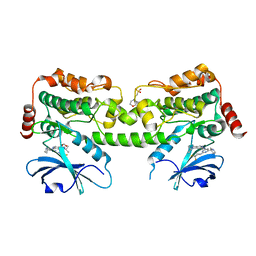 | | Crystal structure of HPK1 in complex with GNE1858 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hematopoietic progenitor kinase, ... | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
6Y6Z
 
 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 1 | | Descriptor: | GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI, ~{tert}-butyl ~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]carbamate | | Authors: | Newman, H, Bellini, D, Dowson, C.G. | | Deposit date: | 2020-02-27 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion.
Chem Sci, 11, 2020
|
|
6Y6O
 
 | | Structure of mature activin A with small molecule 42 | | Descriptor: | (3~{R})-4-ethyl-3-methyl-3-propyl-1~{H}-1,4-benzodiazepine-2,5-dione, Inhibin beta A chain, SULFATE ION | | Authors: | McLoughlin, J.D, Brear, P.B, Reinhardt, T, Spring, D.R, Hyvonen, M. | | Deposit date: | 2020-02-26 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion
Chem Sci, 11, 2020
|
|
6Y6N
 
 | | Structure of mature activin A with small molecule 2 | | Descriptor: | (3~{R})-3,4-dimethyl-3-propyl-1~{H}-1,4-benzodiazepine-2,5-dione, Inhibin beta A chain, SULFATE ION | | Authors: | McLoughlin, J.D, Brear, P.B, Reinhardt, T, Spring, D.R, Hyvonen, M. | | Deposit date: | 2020-02-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion
Chem Sci, 11, 2020
|
|
6Y6U
 
 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 6 | | Descriptor: | 2-(4-hydroxyphenyl)-~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]ethanamide, GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Bellini, D, Dowson, C.G. | | Deposit date: | 2020-02-27 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion
Chem Sci, 11, 2020
|
|
