5WL0
 
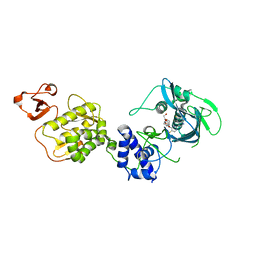 | | Co-crystal structure of Influenza A H3N2 PB2 (241-741) bound to VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, Polymerase basic protein 2 | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2017-07-25 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for therapeutic inhibition of influenza A polymerase PB2 subunit.
Sci Rep, 7, 2017
|
|
7WSO
 
 | | Structure of a membrane protein G | | Descriptor: | B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, Immunoglobulin heavy constant gamma 1 | | Authors: | Ma, X, Zhu, Y, Chen, Y, Huang, Z. | | Deposit date: | 2022-01-30 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM structures of two human B cell receptor isotypes.
Science, 377, 2022
|
|
7XT6
 
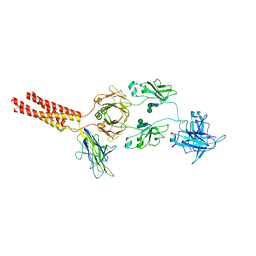 | | Structure of a membrane protein M3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell antigen receptor complex-associated protein alpha chain, ... | | Authors: | Ma, X, Zhu, Y, Chen, Y, Huang, Z. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cryo-EM structures of two human B cell receptor isotypes.
Science, 377, 2022
|
|
7WSP
 
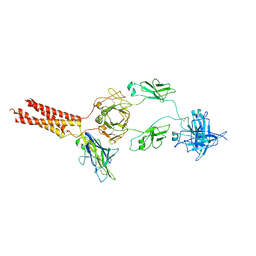 | | Structure of a membrane protein M | | Descriptor: | B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, Isoform 2 of Immunoglobulin heavy constant mu | | Authors: | Ma, X, Zhu, Y, Chen, Y, Huang, Z. | | Deposit date: | 2022-01-30 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Structure of a membrane protein M
To Be Published
|
|
5Z9R
 
 | |
6P9S
 
 | | E.coli LpxA in complex with UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc and Compound 7 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Ma, X, Shia, S, Ornelas, E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two Distinct Mechanisms of Inhibition of LpxA Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
J.Am.Chem.Soc., 142, 2020
|
|
6P9Q
 
 | | E.coli LpxA in complex with UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc and Compound 2 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Ma, X, Shia, S, Ornelas, E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two Distinct Mechanisms of Inhibition of LpxA Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
J.Am.Chem.Soc., 142, 2020
|
|
6P9R
 
 | | E.coli LpxA in complex with UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc and Compound 6 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Ma, X, Shia, S, Ornelas, E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Two Distinct Mechanisms of Inhibition of LpxA Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
J.Am.Chem.Soc., 142, 2020
|
|
6P9P
 
 | | E.coli LpxA in complex with Compound 1 | | Descriptor: | 3-[2-(4-methoxyphenyl)-2-oxoethyl]-5,5-diphenylimidazolidine-2,4-dione, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, ... | | Authors: | Ma, X, Shia, S, Ornelas, E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Distinct Mechanisms of Inhibition of LpxA Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
J.Am.Chem.Soc., 142, 2020
|
|
6P9T
 
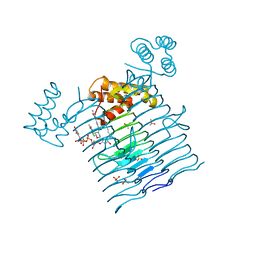 | | E.coli LpxA in complex with UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc and Compound 8 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Ma, X, Shia, S, Ornelas, E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Two Distinct Mechanisms of Inhibition of LpxA Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
J.Am.Chem.Soc., 142, 2020
|
|
6Q2A
 
 | |
3TMP
 
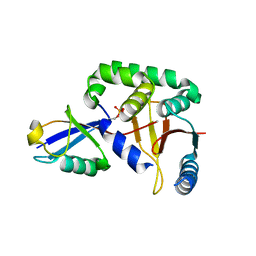 | | The catalytic domain of human deubiquitinase DUBA in complex with ubiquitin aldehyde | | Descriptor: | OTU domain-containing protein 5, Polyubiquitin-C | | Authors: | Ma, X, Yin, J, Hymowitz, S, Starovasnik, M, Cochran, A. | | Deposit date: | 2011-08-31 | | Release date: | 2012-01-11 | | Last modified: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Phosphorylation-dependent activity of the deubiquitinase DUBA.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5EF9
 
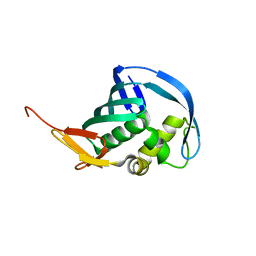 | | Structure of Influenza B Lee PB2 cap-binding domain | | Descriptor: | Polymerase basic protein 2 | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2015-10-23 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis of mRNA Cap Recognition by Influenza B Polymerase PB2 Subunit.
J.Biol.Chem., 291, 2016
|
|
5EFA
 
 | |
5EFC
 
 | |
5BVE
 
 | | Tetrahydropyrrolo-diazepenones as inhibitors of ERK2 kinase | | Descriptor: | 2-[(1S)-1-(3-chlorophenyl)-2-hydroxyethyl]-8-[2-(tetrahydro-2H-pyran-4-ylamino)pyrimidin-4-yl]-2,3,4,5-tetrahydro-1H-pyrrolo[1,2-a][1,4]diazepin-1-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Ma, X, Steven, S. | | Deposit date: | 2015-06-05 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tetrahydropyrrolo-diazepenones as inhibitors of ERK2 kinase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5BVD
 
 | | Tetrahydropyrrolo-diazepenones as inhibitors of ERK2 kinase | | Descriptor: | 2-[(1S)-1-(3-chlorophenyl)-2-hydroxyethyl]-7-[2-(tetrahydro-2H-pyran-4-ylamino)pyrimidin-4-yl]-3,4-dihydropyrrolo[1,2-a]pyrazin-1(2H)-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Ma, X, Steven, S. | | Deposit date: | 2015-06-05 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrahydropyrrolo-diazepenones as inhibitors of ERK2 kinase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5BVF
 
 | | Tetrahydropyrrolo-diazepenones as inhibitors of ERK2 kinase | | Descriptor: | 2-[(1S)-1-(3-chlorophenyl)-2-hydroxyethyl]-8-(2-{[(1S,3R)-3-hydroxycyclopentyl]amino}pyrimidin-4-yl)-2,3,4,5-tetrahydro-1H-pyrrolo[1,2-a][1,4]diazepin-1-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Ma, X, Steven, S. | | Deposit date: | 2015-06-05 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrahydropyrrolo-diazepenones as inhibitors of ERK2 kinase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2P08
 
 | |
2P04
 
 | |
4K3J
 
 | | Crystal structure of Onartuzumab Fab in complex with MET and HGF-beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor beta chain, ... | | Authors: | Ma, X, Starovasnik, M.A. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Monovalent antibody design and mechanism of action of onartuzumab, a MET antagonist with anti-tumor activity as a therapeutic agent.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8DEY
 
 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2,3) and the molecular glue DKY709 | | Descriptor: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Ma, X, Ornelas, E, Clifton, M.C. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
8U17
 
 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-long bound to Pomalidomide | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
8U15
 
 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-short bound to CC-220 | | Descriptor: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DDB1, Protein cereblon, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
8U16
 
 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-short bound to Pomalidomide | | Descriptor: | 1,2-ETHANEDIOL, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
