3OB4
 
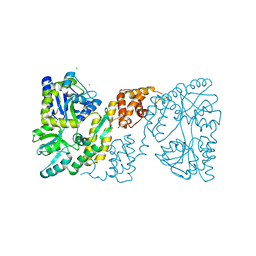 | | MBP-fusion protein of the major peanut allergen Ara h 2 | | Descriptor: | CHLORIDE ION, Maltose ABC transporter periplasmic protein,Arah 2, SULFATE ION, ... | | Authors: | Mueller, G.A, Gosavi, R.A, Moon, A.F, London, R.E, Pedersen, L.C. | | Deposit date: | 2010-08-06 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Ara h 2: crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity.
Allergy, 66, 2011
|
|
3OWV
 
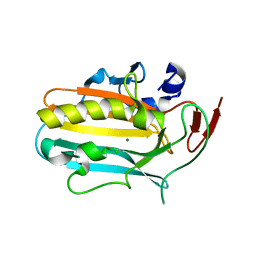 | | Structural insights into catalytic and substrate binding mechanisms of the strategic EndA nuclease from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, DNA-entry nuclease, MAGNESIUM ION | | Authors: | Moon, A.F, Midon, M, Meiss, G, Pingoud, A.M, London, R.E, Pedersen, L.C. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into catalytic and substrate binding mechanisms of the strategic EndA nuclease from Streptococcus pneumoniae.
Nucleic Acids Res., 39, 2011
|
|
3PC6
 
 | |
3PC7
 
 | |
3PC8
 
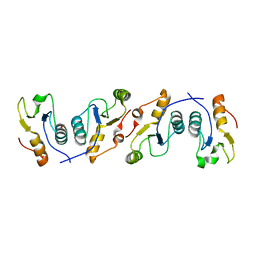 | | X-ray crystal structure of the heterodimeric complex of XRCC1 and DNA ligase III-alpha BRCT domains. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA ligase 3, DNA repair protein XRCC1, ... | | Authors: | Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2010-10-21 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The structural basis for partitioning of the XRCC1/DNA ligase III-{alpha} BRCT-mediated dimer complexes.
Nucleic Acids Res., 39, 2011
|
|
3QVG
 
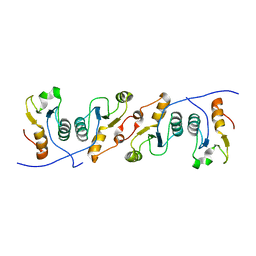 | | XRCC1 bound to DNA ligase | | Descriptor: | DNA ligase 3, DNA repair protein XRCC1 | | Authors: | Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2011-02-25 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The structural basis for partitioning of the XRCC1/DNA ligase III-{alpha} BRCT-mediated dimer complexes.
Nucleic Acids Res., 39, 2011
|
|
5UII
 
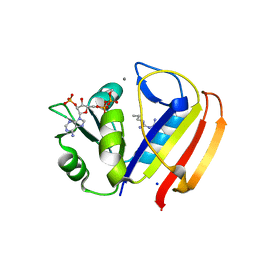 | |
5UIP
 
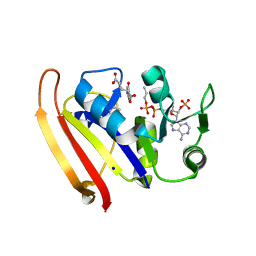 | | structure of DHFR with bound DAP, p-ABG and NADP | | Descriptor: | Dihydrofolate reductase, N-(4-aminobenzene-1-carbonyl)-L-glutamic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pedersen, L.C, London, R.E. | | Deposit date: | 2017-01-14 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Structural Basis for Biguanide Activity.
Biochemistry, 56, 2017
|
|
5UIO
 
 | |
5UIH
 
 | |
5W4F
 
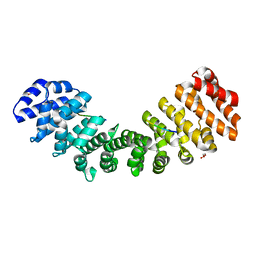 | | Importin binding to pol Mu NLS peptide | | Descriptor: | DNA-directed DNA/RNA polymerase mu, GLYCEROL, Importin subunit alpha-1 | | Authors: | Pedersen, L.C, London, R.E. | | Deposit date: | 2017-06-10 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Variations in nuclear localization strategies among pol X family enzymes.
Traffic, 2018
|
|
5W7Y
 
 | |
5W7W
 
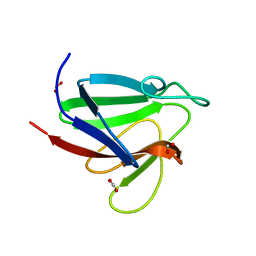 | | Crystal Structure of FHA domain of human APLF | | Descriptor: | Aprataxin and PNK-like factor, FORMIC ACID, SODIUM ION | | Authors: | Pedersen, L.C, Kim, K, London, R.E. | | Deposit date: | 2017-06-21 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.348 Å) | | Cite: | Characterization of the APLF FHA-XRCC1 phosphopeptide interaction and its structural and functional implications.
Nucleic Acids Res., 45, 2017
|
|
5W7X
 
 | |
2L7D
 
 | | Ribonucleotide Perturbation of DNA Structure: Solution Structure of [d(CGC)r(G)d(AATTCGCG)]2 | | Descriptor: | 5'-D(*CP*GP*C)-R(P*G)-D(P*AP*AP*TP*TP*CP*GP*CP*G)-3' | | Authors: | DeRose, E.F, Perera, L, Murray, M.S, Kunkel, T.A, London, R.E. | | Deposit date: | 2010-12-07 | | Release date: | 2011-06-01 | | Last modified: | 2012-06-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Dickerson DNA dodecamer containing a single ribonucleotide.
Biochemistry, 51, 2012
|
|
2MDP
 
 | |
2O3B
 
 | | Crystal structure complex of Nuclease A (NucA) with intra-cellular inhibitor NuiA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Ghosh, M, Meiss, G, Pingoud, A.M, London, R.E, Pedersen, L.C. | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The nuclease a-inhibitor complex is characterized by a novel metal ion bridge.
J.Biol.Chem., 282, 2007
|
|
2RH2
 
 | | High Resolution DHFR R-67 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Dihydrofolate reductase type 2 | | Authors: | Krahn, J.M, London, R.E. | | Deposit date: | 2007-10-05 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Crystal Structure of a Type II Dihydrofolate Reductase Catalytic Ternary Complex
Biochemistry, 46, 2007
|
|
2RK1
 
 | | DHFR R67 Complexed with NADP and dihydrofolate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DIHYDROFOLIC ACID, Dihydrofolate reductase type 2, ... | | Authors: | Krahn, J.M, London, R.E. | | Deposit date: | 2007-10-16 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of a type II dihydrofolate reductase catalytic ternary complex.
Biochemistry, 46, 2007
|
|
2RK2
 
 | | DHFR R-67 complexed with NADP | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Dihydrofolate reductase type 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Krahn, J.M, London, R.E. | | Deposit date: | 2007-10-16 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a type II dihydrofolate reductase catalytic ternary complex.
Biochemistry, 46, 2007
|
|
1J57
 
 | | NuiA | | Descriptor: | NuiA | | Authors: | Kirby, T.W, Mueller, G.A, DeRose, E.F, Lebetkin, M.S, Meiss, G, Pingoud, A, London, R.E. | | Deposit date: | 2002-01-17 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The Nuclease A Inhibitor represents a new variation of the rare PR-1 fold.
J.Mol.Biol., 320, 2002
|
|
1KTU
 
 | | NuiA | | Descriptor: | NuiA | | Authors: | Kirby, T.W, Mueller, G.A, DeRose, E.F, Lebetkin, M.S, Meiss, G, Pingoud, A, London, R.E. | | Deposit date: | 2002-01-17 | | Release date: | 2002-12-04 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The Nuclease A Inhibitor represents a new variation of the rare PR-1 fold.
J.Mol.Biol., 320, 2002
|
|
1NZP
 
 | | Solution Structure of the Lyase Domain of Human DNA Polymerase Lambda | | Descriptor: | DNA polymerase lambda | | Authors: | DeRose, E.F, Kirby, T.W, Mueller, G.A, Bebenek, K, Garcia-Diaz, M, Blanco, L, Kunkel, T.A, London, R.E. | | Deposit date: | 2003-02-19 | | Release date: | 2003-08-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Lyase Domain of Human DNA Polymerase Lambda
Biochemistry, 42, 2003
|
|
1O1W
 
 | | SOLUTION STRUCTURE OF THE RNASE H DOMAIN OF THE HIV-1 REVERSE TRANSCRIPTASE IN THE PRESENCE OF MAGNESIUM | | Descriptor: | RIBONUCLEASE H | | Authors: | Pari, K, Mueller, G.A, Derose, E.F, Kirby, T.W, London, R.E. | | Deposit date: | 2003-02-12 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Rnase H Domain of the HIV-1 Reverse Transcriptase in the Presence of Magnesium
Biochemistry, 42, 2003
|
|
1QVX
 
 | | SOLUTION STRUCTURE OF THE FAT DOMAIN OF FOCAL ADHESION KINASE | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Gao, G, Prutzman, K.C, King, M.L, DeRose, E.F, London, R.E, Schaller, M.D, Campbell, S.L. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Focal Adhesion Targeting Domain of Focal Adhesion Kinase in Complex with a Paxillin LD Peptide: EVIDENCE FOR A TWO-SITE BINDING MODEL.
J.Biol.Chem., 279, 2004
|
|
