2RBH
 
 | | Gamma-glutamyl cyclotransferase | | Descriptor: | Gamma-glutamyl cyclotransferase | | Authors: | Oakley, A.J, Board, P.G. | | Deposit date: | 2007-09-19 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The identification and structural characterization of C7orf24 as gamma-glutamyl cyclotransferase. An essential enzyme in the gamma-glutamyl cycle
J.Biol.Chem., 283, 2008
|
|
7VIJ
 
 | | Crystal structure of USP7-HUBL domain | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Feng, N, Zeng, K.W. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Neuroinflammation inhibition by small-molecule targeting USP7 noncatalytic domain for neurodegenerative disease therapy.
Sci Adv, 8, 2022
|
|
7XPY
 
 | | Crystal structure of USP7 in complex with its inhibitor | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, [(3S,3aR,4R,6Z,9S,10E,11aR)-9-acetyloxy-6-(acetyloxymethyl)-3,10-dimethyl-2-oxidanylidene-3a,4,5,8,9,11a-hexahydro-3H-cyclodeca[b]furan-4-yl] (E)-2-methyl-4-oxidanyl-but-2-enoate | | Authors: | Feng, N, Zeng, K.W. | | Deposit date: | 2022-05-06 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Neuroinflammation inhibition by small-molecule targeting USP7 noncatalytic domain for neurodegenerative disease therapy.
Sci Adv, 8, 2022
|
|
6J20
 
 | | Crystal structure of the human NK1 substance P receptor | | Descriptor: | 5-[[(2~{R},3~{S})-2-[(1~{R})-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy]-3-(4-fluorophenyl)morpholin-4-yl]methyl]-1,2-dihydro-1,2,4-triazol-3-one, Substance-P receptor,Endolysin | | Authors: | Chen, S, Lu, M, Zhang, H, Wu, B, Zhao, Q. | | Deposit date: | 2018-12-30 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography.
Nat Commun, 10, 2019
|
|
6J21
 
 | | Crystal structure of the human NK1 substance P receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-[[(2~{R},3~{S})-2-[(1~{R})-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy]-3-(4-fluorophenyl)morpholin-4-yl]methyl]-1,2-dihydro-1,2,4-triazol-3-one, Substance-P receptor,Endolysin | | Authors: | Chen, S, Lu, M, Zhang, H, Wu, B, Zhao, Q. | | Deposit date: | 2018-12-30 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography.
Nat Commun, 10, 2019
|
|
6L6Z
 
 | |
6LFG
 
 | |
2G6U
 
 | |
3Q19
 
 | | Human Glutathione Transferase O2 | | Descriptor: | CHLORIDE ION, GLUTATHIONE, Glutathione S-transferase omega-2 | | Authors: | Zhou, H, Board, P.G, Oakley, A.J. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the dehydroascorbate reductase activity of human omega-class glutathione transferases.
J.Mol.Biol., 420, 2012
|
|
3Q18
 
 | | Human Glutathione Transferase O2 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zhou, H, Board, P.G, Oakley, A.J. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the dehydroascorbate reductase activity of human omega-class glutathione transferases.
J.Mol.Biol., 420, 2012
|
|
3QAG
 
 | | Human Glutathione Transferase O2 with glutathione -new crystal form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Zhou, H, Board, P.G, Oakley, A.J. | | Deposit date: | 2011-01-11 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the dehydroascorbate reductase activity of human omega-class glutathione transferases.
J.Mol.Biol., 420, 2012
|
|
8JBG
 
 | | Neurokinin B bound to active human neurokinin 3 receptor in complex with Gq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein Gq (G324), ... | | Authors: | Sun, W.J, Yang, F, Zhang, H.H, Yuan, Q.N, Yin, W.C, Shi, P, Eric, X, Tian, C.L. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into neurokinin 3 receptor activation by endogenous and analogue peptide agonists.
Cell Discov, 9, 2023
|
|
8JBH
 
 | | Substance P bound to active human neurokinin 3 receptor in complex with Gq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein Gq subunit alpha (G324), ... | | Authors: | Sun, W.J, Yang, F, Zhang, H.H, Yuan, Q.N, Yin, W.C, Shi, P, Eric, X, Tian, C.L. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into neurokinin 3 receptor activation by endogenous and analogue peptide agonists.
Cell Discov, 9, 2023
|
|
8JBF
 
 | | Senktide bound to active human neurokinin 3 receptor in complex with Gq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein Gq subunit alpha, ... | | Authors: | Sun, W.J, Yang, F, Zhang, H.H, Yuan, Q.N, Yin, W.C, Shi, P, Eric, X, Tian, C.L. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into neurokinin 3 receptor activation by endogenous and analogue peptide agonists.
Cell Discov, 9, 2023
|
|
3RZ2
 
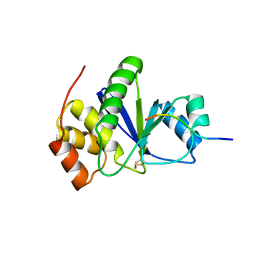 | | Crystal of Prl-1 complexed with peptide | | Descriptor: | Prl-1 (PTP4A1), Protein tyrosine phosphatase type IVA 1 | | Authors: | Zhang, Z.-Y, Liu, D, Bai, Y. | | Deposit date: | 2011-05-11 | | Release date: | 2011-10-26 | | Last modified: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | PRL-1 protein promotes ERK1/2 and RhoA protein activation through a non-canonical interaction with the Src homology 3 domain of p115 Rho GTPase-activating protein.
J.Biol.Chem., 286, 2011
|
|
5T1S
 
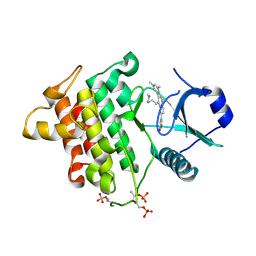 | | Irak4 kinase - compound 1 co-structure | | Descriptor: | 5-[3-(3,5-dimethylphenyl)-4-[4-(methylamino)butyl]quinolin-6-yl]pyridin-3-ol, Interleukin-1 receptor-associated kinase 4 | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of quinazoline based inhibitors of IRAK4 for the treatment of inflammation.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5T1T
 
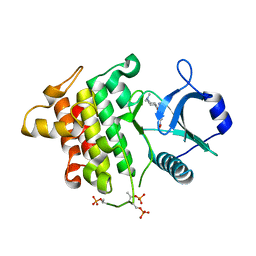 | | Irak4 kinase - compound 1 co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N},~{N}-dimethyl-4-(6-nitroquinazolin-4-yl)oxy-cyclohexan-1-amine | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Identification of quinazoline based inhibitors of IRAK4 for the treatment of inflammation.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5W4Q
 
 | |
5W4P
 
 | |
5W4O
 
 | |
5WO4
 
 | | JAK1 complexed with compound 28 | | Descriptor: | 3-[(4-chloro-3-methoxyphenyl)amino]-1-[(3R,4S)-4-cyanooxan-3-yl]-1H-pyrazole-4-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lesburg, C.A, Patel, S.B. | | Deposit date: | 2017-08-01 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Discovery of 3-((4-Chloro-3-methoxyphenyl)amino)-1-((3R,4S)-4-cyanotetrahydro-2H-pyran-3-yl)-1H-pyrazole-4-carboxamide, a Highly Ligand Efficient and Efficacious Janus Kinase 1 Selective Inhibitor with Favorable Pharmacokinetic Properties.
J. Med. Chem., 60, 2017
|
|
7XQ8
 
 | | Structure of human B-cell antigen receptor of the IgM isotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, ... | | Authors: | Chen, M.Y, Su, Q, Shi, Y.G. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-17 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the human IgM B cell receptor.
Science, 377, 2022
|
|
3UIM
 
 | |
3VLN
 
 | | Human Glutathione Transferase O1-1 C32S Mutant in Complex with Ascorbic Acid | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ASCORBIC ACID, ... | | Authors: | Brock, J, Board, P.G, Oakley, A.J. | | Deposit date: | 2011-12-02 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the dehydroascorbate reductase activity of human omega-class glutathione transferases.
J.Mol.Biol., 420, 2012
|
|
6AFB
 
 | | DJ-1 C106S incubated with isatin | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, ISATIN, ... | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
