4XSJ
 
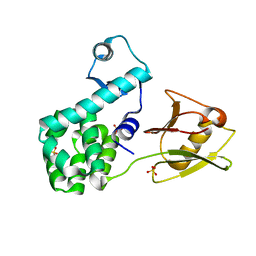 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
4XTB
 
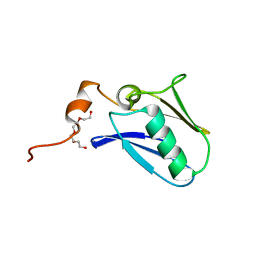 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter | | Descriptor: | Calcium uniporter protein, mitochondrial, TETRAETHYLENE GLYCOL | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
5BZ6
 
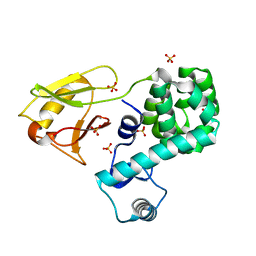 | | Crystal structure of the N-terminal domain single mutant (S92A) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-06-11 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
6GS4
 
 | | Crystal structure of peptide transporter DtpA-nanobody in complex with valganciclovir | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Dipeptide and tripeptide permease A, [(2~{S})-2-[(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)methoxy]-3-oxidanyl-propyl] (2~{S})-2-azanyl-3-methyl-butanoate, ... | | Authors: | Ural-Blimke, Y, Flayhan, A, Quistgaard, E.M, Loew, C. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
6GS7
 
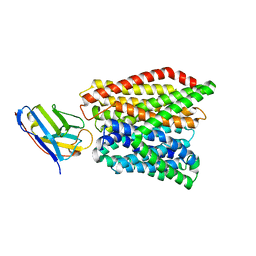 | | Crystal structure of peptide transporter DtpA-nanobody in glycine buffer | | Descriptor: | Dipeptide and tripeptide permease A, nanobody | | Authors: | Ural-Blimke, Y, Flayhan, A, Quistgaard, E.M, Loew, C. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
6GS1
 
 | | Crystal structure of peptide transporter DtpA-nanobody in MES buffer | | Descriptor: | Dipeptide and tripeptide permease A, Nanobody 00 | | Authors: | Ural-Blimke, Y, Flayhan, A, Loew, C, Quistgaard, E.M. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
4UV7
 
 | | The complex structure of extracellular domain of EGFR and GC1118A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Yoo, J.H, Cho, H.S. | | Deposit date: | 2014-08-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gc1118, an Anti-Egfr Antibody with a Distinct Binding Epitope and Superior Inhibitory Activity Against High-Affinity Egfr Ligands.
Mol.Cancer Ther., 15, 2016
|
|
5D50
 
 | | Crystal structure of Rep-Ant complex from Salmonella-temperate phage | | Descriptor: | Anti-repressor protein, Repressor | | Authors: | Son, S.H, Yoon, H.J, Ryu, S, Lee, H.H. | | Deposit date: | 2015-08-10 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Noncanonical DNA-binding mode of repressor and its disassembly by antirepressor
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3FVH
 
 | |
3C5L
 
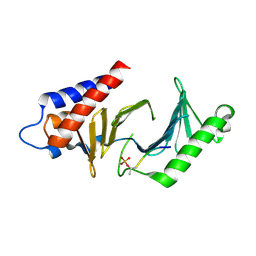 | |
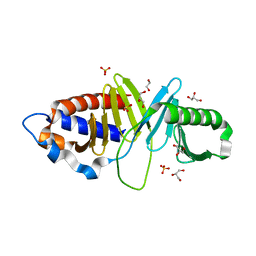
3HIH
 
 | |
3HIK
 
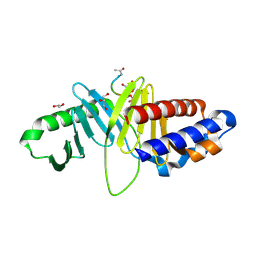 | | Structure of human Plk1-PBD in complex with PLHSpT | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pentamer phosphopeptide, ... | | Authors: | Wlodawer, A, Moulaei, T. | | Deposit date: | 2009-05-20 | | Release date: | 2009-06-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and functional analyses of minimal phosphopeptides targeting the polo-box domain of polo-like kinase 1.
Nat.Struct.Mol.Biol., 16, 2009
|
|
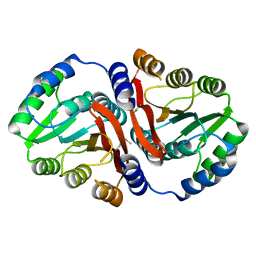
3DUL
 
 | |
3DUW
 
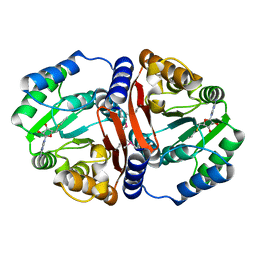 | |
5D4Z
 
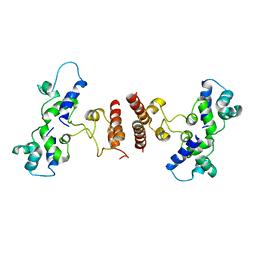 | |
1EJP
 
 | | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN | | Descriptor: | SYNDECAN-4 | | Authors: | Lee, D, Oh, E.S, Woods, A, Couchman, J.R, Lee, W. | | Deposit date: | 2000-03-03 | | Release date: | 2001-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dimeric cytoplasmic domain of syndecan-4.
Biochemistry, 40, 2001
|
|
4ODP
 
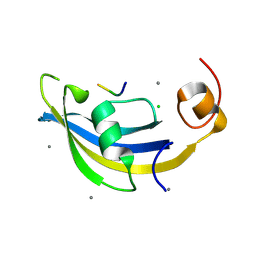 | | Structure of SlyD delta-IF from Thermus thermophilus in complex with S2-W23A peptide | | Descriptor: | 30S ribosomal protein S2, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Quistgaard, E.M, Low, C, Nordlund, P. | | Deposit date: | 2014-01-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD.
BMC Biol., 14, 2016
|
|
4ODO
 
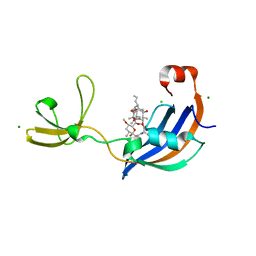 | | Structure of SlyD from Thermus thermophilus in complex with FK506 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CHLORIDE ION, ... | | Authors: | Quistgaard, E.M, Low, C, Nordlund, P. | | Deposit date: | 2014-01-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD.
BMC Biol., 14, 2016
|
|
4ODM
 
 | | Structure of SlyD from Thermus thermophilus in complex with S2-W23A peptide | | Descriptor: | 30S ribosomal protein S2, ACETATE ION, CHLORIDE ION, ... | | Authors: | Quistgaard, E.M, Low, C, Nordlund, P. | | Deposit date: | 2014-01-10 | | Release date: | 2015-01-14 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD.
BMC Biol., 14, 2016
|
|
4ODK
 
 | |
4ODR
 
 | | Structure of SlyD delta-IF from Thermus thermophilus in complex with FK506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, ACETATE ION, CHLORIDE ION, ... | | Authors: | Quistgaard, E.M, Low, C, Nordlund, P. | | Deposit date: | 2014-01-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD.
BMC Biol., 14, 2016
|
|
4ODQ
 
 | | Structure of SlyD delta-IF from Thermus thermophilus in complex with S3 peptide | | Descriptor: | 30S ribosomal protein S3, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Quistgaard, E.M, Low, C, Nordlund, P. | | Deposit date: | 2014-01-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD.
BMC Biol., 14, 2016
|
|
4ODN
 
 | | Structure of SlyD from Thermus thermophilus in complex with S2-plus peptide | | Descriptor: | 30S ribosomal protein S2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Quistgaard, E.M, Low, C, Nordlund, P. | | Deposit date: | 2014-01-10 | | Release date: | 2015-01-14 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD.
BMC Biol., 14, 2016
|
|
4ODL
 
 | |
3CHM
 
 | |
