2P6B
 
 | | Crystal Structure of Human Calcineurin in Complex with PVIVIT Peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B isoform 1, Calmodulin-dependent calcineurin A subunit alpha isoform, ... | | Authors: | Li, H, Zhang, L, Rao, A, Harrison, S.C, Hogan, P.G. | | Deposit date: | 2007-03-16 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of calcineurin in complex with PVIVIT peptide: Portrait of a low-affinity signalling interaction
J.Mol.Biol., 369, 2007
|
|
3MD1
 
 | | Crystal Structure of the Second RRM Domain of Yeast Poly(U)-Binding Protein (Pub1) | | Descriptor: | GLYCEROL, Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1 | | Authors: | Li, H, Shi, H, Li, Y, Cui, Y, Niu, L, Teng, M. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Second RRM Domain of Yeast Poly(U)-Binding Protein (Pub1)
To be published
|
|
9NSE
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, ETHYL-ISOSELENOUREA COMPLEX | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLIC ACID, ... | | Authors: | Li, H, Raman, C.S, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-01-13 | | Release date: | 2000-10-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Mapping the active site polarity in structures of endothelial nitric oxide synthase heme domain complexed with isothioureas.
J.Inorg.Biochem., 81, 2000
|
|
5K1A
 
 | |
5K1C
 
 | | Crystal structure of the UAF1/WDR20/USP12 complex | | Descriptor: | PHOSPHATE ION, TRIS(HYDROXYETHYL)AMINOMETHANE, Ubiquitin carboxyl-terminal hydrolase 12, ... | | Authors: | Li, H, D'Andrea, A.D, Zheng, N. | | Deposit date: | 2016-05-18 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Allosteric Activation of Ubiquitin-Specific Proteases by beta-Propeller Proteins UAF1 and WDR20.
Mol.Cell, 63, 2016
|
|
5K16
 
 | |
5K1B
 
 | |
5E6F
 
 | | Canarypox virus resolvase | | Descriptor: | CNPV261 Holliday junction resolvase protein, D(-)-TARTARIC ACID, MAGNESIUM ION | | Authors: | Li, H, Hwang, Y, Perry, K, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Metal Binding Properties of a Poxvirus Resolvase.
J.Biol.Chem., 291, 2016
|
|
4G3U
 
 | |
2E6J
 
 | | Solution structure of the C-terminal PapD-like domain from human HYDIN protein | | Descriptor: | HYDIN protein | | Authors: | Li, H, Sato, M, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-27 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal PapD-like domain from human HYDIN protein
To be Published
|
|
2E6I
 
 | | Solution structure of the BTK motif of tyrosine-protein kinase ITK from human | | Descriptor: | Tyrosine-protein kinase ITK/TSK, ZINC ION | | Authors: | Li, H, Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-27 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the BTK motif of tyrosine-protein kinase ITK from human
To be Published
|
|
8SGK
 
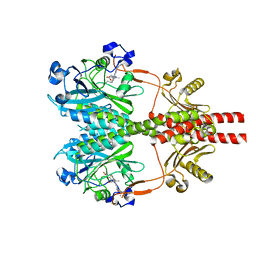 | | CryoEM structure of Deinococcus radiodurans BphP photosensory module in Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Li, H, Li, H. | | Deposit date: | 2023-04-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CryoEM structure of Deinococcus radiodurans BphP photosensory module in Pr state
To Be Published
|
|
7SCK
 
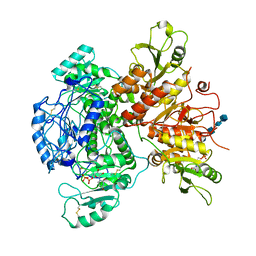 | |
7SCJ
 
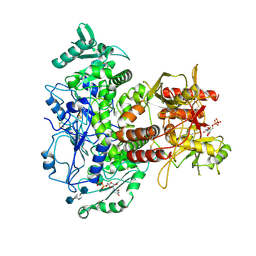 | |
7SCH
 
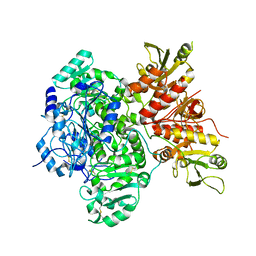 | | Cryo-EM structure of the human Exostosin-1 and Exostosin-2 heterodimer | | Descriptor: | Exostosin-1, Exostosin-2, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, H, Li, H. | | Deposit date: | 2021-09-28 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for heparan sulfate co-polymerase action by the EXT1-2 complex.
Nat.Chem.Biol., 19, 2023
|
|
3P5T
 
 | | CFIm25-CFIm68 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Li, H, Tong, S, Li, X, Shi, H, Gao, Y, Ge, H, Niu, L, Teng, M. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of pre-mRNA recognition by the human cleavage factor Im complex
To be Published
|
|
3P6Y
 
 | | CF Im25-CF Im68-UGUAA complex | | Descriptor: | 5'-R(*UP*GP*UP*AP*A)-3', Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Li, H, Tong, S, Li, X, Shi, H, Gao, Y, Ge, H, Niu, L, Teng, M. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of pre-mRNA recognition by the human cleavage factor Im complex
To be Published
|
|
7S05
 
 | | Cryo-EM structure of human GlcNAc-1-phosphotransferase A2B2 subcomplex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, H, Li, H. | | Deposit date: | 2021-08-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human GlcNAc-1-phosphotransferase alpha beta subunits reveals regulatory mechanism for lysosomal enzyme glycan phosphorylation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S06
 
 | | Cryo-EM structure of human GlcNAc-1-phosphotransferase A2B2 subcomplex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosamine-1-phosphotransferase subunits alpha/beta | | Authors: | Li, H, Li, H. | | Deposit date: | 2021-08-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human GlcNAc-1-phosphotransferase alpha beta subunits reveals regulatory mechanism for lysosomal enzyme glycan phosphorylation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RZW
 
 | | CryoEM structure of Arabidopsis thaliana phytochrome B | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B | | Authors: | Li, H, Burgie, E.S, Vierstra, R.D, Li, H. | | Deposit date: | 2021-08-27 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Plant phytochrome B is an asymmetric dimer with unique signalling potential.
Nature, 604, 2022
|
|
1LZZ
 
 | | Rat neuronal NOS heme domain with N-isopropyl-N'-hydroxyguanidine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-ISOPROPYL-N'-HYDROXYGUANIDINE, ... | | Authors: | Li, H, Shimizu, H, Flinspach, M, Jamal, J, Yang, W, Xian, M, Cai, T, Wen, E.Z, Jia, Q, Wang, P.G, Poulos, T.L. | | Deposit date: | 2002-06-11 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Novel Binding Mode of N-Alkyl-N'-Hydroxyguanidine to Neuronal Nitric Oxide
Synthase Provides Mechanistic Insights into NO Biosynthesis
Biochemistry, 41, 2002
|
|
1LZX
 
 | | Rat neuronal NOS heme domain with NG-hydroxy-L-arginine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-OMEGA-HYDROXY-L-ARGININE, ... | | Authors: | Li, H, Shimizu, H, Flinspach, M, Jamal, J, Yang, W, Xian, M, Cai, T, Wen, E.Z, Jia, Q, Wang, P.G, Poulos, T.L. | | Deposit date: | 2002-06-11 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Novel Binding Mode of N-Alkyl-N'-Hydroxyguanidine to Neuronal Nitric Oxide
Synthase Provides Mechanistic Insights into NO Biosynthesis
Biochemistry, 41, 2002
|
|
1ADC
 
 | | CRYSTALLOGRAPHIC STUDIES OF ISOSTERIC NAD ANALOGUES BOUND TO ALCOHOL DEHYDROGENASE: SPECIFICITY AND SUBSTRATE BINDING IN TWO TERNARY COMPLEXES | | Descriptor: | 5-BETA-D-RIBOFURANOSYLPICOLINAMIDE ADENINE-DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, ETHANOL, ... | | Authors: | Li, H, Hallows, W.A, Punzi, J.S, Pankiewicz, K.W, Watanabe, K.A, Goldstein, B.M. | | Deposit date: | 1993-12-13 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of isosteric NAD analogues bound to alcohol dehydrogenase: specificity and substrate binding in two ternary complexes.
Biochemistry, 33, 1994
|
|
2AXJ
 
 | | Crystal structures of T cell receptor beta chains related to rheumatoid arthritis | | Descriptor: | SF4 T cell receptor beta chain | | Authors: | Li, H, Van Vranken, S, Zhao, Y, Li, Z, Guo, Y, Eisele, L, Li, Y. | | Deposit date: | 2005-09-05 | | Release date: | 2005-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of T cell receptor (beta) chains related to rheumatoid arthritis.
Protein Sci., 14, 2005
|
|
2AXH
 
 | | Crystal structures of T cell receptor beta chains related to rheumatoid arthritis | | Descriptor: | T cell receptor beta chain | | Authors: | Li, H, Van Vranken, S, Zhao, Y, Li, Z, Guo, Y, Eisele, L, Li, Y. | | Deposit date: | 2005-09-05 | | Release date: | 2005-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of T cell receptor (beta) chains related to rheumatoid arthritis.
Protein Sci., 14, 2005
|
|
