4BGQ
 
 | | Crystal structure of the human CDKL5 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CYCLIN-DEPENDENT KINASE-LIKE 5, ... | | Authors: | Canning, P, Krojer, T, Goubin, S, Mahajan, P, Vollmar, M, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CDKL Family Kinases Have Evolved Distinct Structural Features and Ciliary Function.
Cell Rep, 22, 2018
|
|
7PUJ
 
 | | Crystal structure of Endoglycosidase E GH18 domain from Enterococcus faecalis | | Descriptor: | Beta-N-acetylhexosaminidase, CHLORIDE ION, ZINC ION | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
7PUL
 
 | | Crystal structure of Endoglycosidase E GH20 domain from Enterococcus faecalis | | Descriptor: | Beta-N-acetylhexosaminidase, CALCIUM ION, GLY-ALA-GLY-ALA-ALA, ... | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
7PUK
 
 | | Crystal structure of Endoglycosidase E GH18 domain from Enterococcus faecalis in complex with Man5 product | | Descriptor: | Beta-N-acetylhexosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
3QAH
 
 | | Crystal structure of apo-form human MOF catalytic domain | | Descriptor: | Probable histone acetyltransferase MYST1, ZINC ION | | Authors: | Sun, B, Tang, Q, Zhong, C, Ding, J. | | Deposit date: | 2011-01-11 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regulation of the histone acetyltransferase activity of hMOF via autoacetylation of Lys274
Cell Res., 21, 2011
|
|
6AG0
 
 | | The X-ray Crystallographic Structure of Maltooligosaccharide-forming Amylase from Bacillus stearothermophilus STB04 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase, CALCIUM ION | | Authors: | Li, Z.F, Li, Y.L, Ban, X.F, Zhang, C.Y, Jin, T.C, Xie, X.F, Gu, Z.B, Li, C.M. | | Deposit date: | 2018-08-09 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a maltooligosaccharide-forming amylase from Bacillus stearothermophilus STB04.
Int.J.Biol.Macromol., 138, 2019
|
|
4YVP
 
 | | Crystal Structure of AKR1C1 complexed with glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4ZFC
 
 | | Crystal structure of AKR1C3 complexed with glicazide | | Descriptor: | Aldo-keto reductase family 1 member C3, N-[(3aR,6aS)-hexahydrocyclopenta[c]pyrrol-2(1H)-ylcarbamoyl]-4-methylbenzenesulfonamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zhrng, X, Hu, X. | | Deposit date: | 2015-04-21 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
2HC1
 
 | | Engineered catalytic domain of protein tyrosine phosphatase HPTPbeta. | | Descriptor: | ACETATE ION, CHLORIDE ION, Receptor-type tyrosine-protein phosphatase beta | | Authors: | Evdokimov, A.G, Pokross, M, Walter, R, Mekel, M. | | Deposit date: | 2006-06-14 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2HC2
 
 | | Engineered protein tyrosine phosphatase beta catalytic domain | | Descriptor: | MAGNESIUM ION, Receptor-type tyrosine-protein phosphatase beta, SODIUM ION | | Authors: | Evdokimov, A.G, Pokross, M, Walter, R, Mekel, M. | | Deposit date: | 2006-06-15 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5Y93
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[5-[(5-bromanyl-2-methoxy-phenyl)sulfonylamino]-3-methyl-1,2-benzoxazol-6-yl]oxy]-N-(2-morpholin-4-ylethyl)ethanamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y8W
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-N-(3-methyl-6-oxidanyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y94
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 5-bromanyl-2-methoxy-N-[3-methyl-6-(methylamino)-1,2-benzoxazol-5-yl]benzenesulfonamide, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y8Z
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-N-(3,6-dimethyl-1,2-benzoxazol-5-yl)-2-methoxy-benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y8Y
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y8C
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloranyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
3L9M
 
 | | Crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 18 | | Descriptor: | (2S)-N~1~-[5-(3-methyl-1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L9N
 
 | | crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 27 | | Descriptor: | (2S)-N~1~-[5-(1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L9L
 
 | | Crystal structure of pka with compound 36 | | Descriptor: | 5-[2-({(2S)-2-amino-3-[4-(trifluoromethyl)phenyl]propyl}amino)-1,3-thiazol-5-yl]-1,3-dihydro-2H-indol-2-one, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4PPI
 
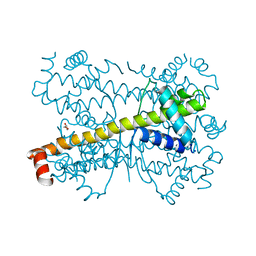 | | Crystal structure of Bcl-xL hexamer | | Descriptor: | Bcl-2-like protein 1, GLYCEROL | | Authors: | Sreekanth, R, Yoon, H.S. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Structural transition in Bcl-xL and its potential association with mitochondrial calcium ion transport
Sci Rep, 5, 2015
|
|
4QBQ
 
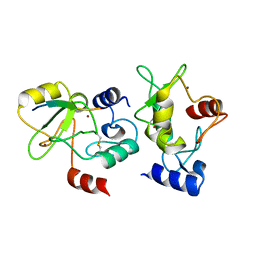 | | Crystal structure of DNMT3a ADD domain bound to H3 peptide | | Descriptor: | DNA (cytosine-5)-methyltransferase 3A, Histone H3, ZINC ION | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2014-05-08 | | Release date: | 2015-05-13 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Engineering of a histone-recognition domain in a de novo DNA methyltransferase alters the epigenetic landscape of ESCs
To be Published
|
|
4QBS
 
 | |
4QBR
 
 | |
3NSQ
 
 | | Crystal structure of tetrameric RXRalpha-LBD complexed with antagonist danthron | | Descriptor: | 1,8-dihydroxyanthracene-9,10-dione, Retinoid X receptor, alpha | | Authors: | Zhang, H, Hu, T, Li, L, Zhou, R, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor.
J.Biol.Chem., 286, 2011
|
|
6IUO
 
 | | Crystal structure of FGFR4 kinase domain in complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-({4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}methyl)prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
