3AQV
 
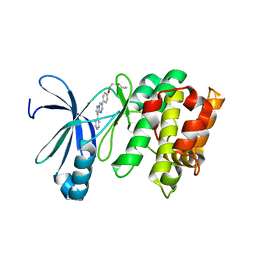 | | Human AMP-activated protein kinase alpha 2 subunit kinase domain (T172D) complexed with compound C | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine | | Authors: | Handa, N, Takagi, T, Saijo, S, Kishishita, S, Toyama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for compound C inhibition of the human AMP-activated protein kinase alpha 2 subunit kinase domain
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5A34
 
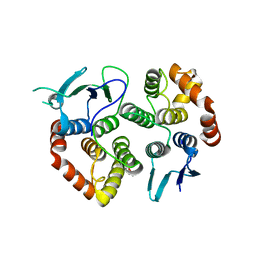 | | The crystal structure of the GST-like domains complex of EPRS-AIMP2 | | Descriptor: | AMINOACYL TRNA SYNTHASE COMPLEX-INTERACTING MULTIFUNCTIONAL PROTEIN 2, BIFUNCTIONAL GLUTAMATE/PROLINE--TRNA LIGASE, GLYCEROL | | Authors: | Cho, H.Y, Kang, B.S. | | Deposit date: | 2015-05-27 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Assembly of Multi-tRNA Synthetase Complex Via Heterotetrameric Glutathione Transferase-Homology Domains.
J.Biol.Chem., 290, 2015
|
|
8GSV
 
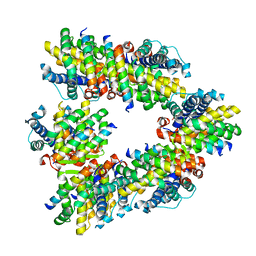 | | Crystal structure of human BAK in complex with the Pxt1 BH3 domain | | Descriptor: | Bcl-2 homologous antagonist/killer, Peroxisomal testis-specific protein 1 | | Authors: | Lim, D, Ku, B. | | Deposit date: | 2022-09-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for proapoptotic activation of Bak by the noncanonical BH3-only protein Pxt1.
Plos Biol., 21, 2023
|
|
5XJG
 
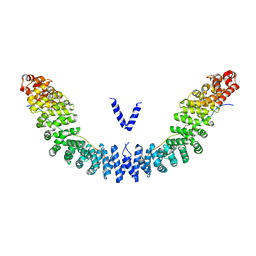 | | Crystal structure of Vac8p bound to Nvj1p | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Nucleus-vacuole junction protein 1, ... | | Authors: | Jeong, H, Park, J, Jun, Y, Lee, C. | | Deposit date: | 2017-05-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insight into the nucleus-vacuole junction based on the Vac8p-Nvj1p crystal structure.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3V68
 
 | |
6KNR
 
 | | Crystal structure of Estrogen-related receptor gamma ligand-binding domain with DN200699 | | Descriptor: | (E)-4-(1-(4-(1-cyclopropylpiperidin-4-yl)phenyl)-5-hydroxy-2-phenylpent-1-en-1-yl)phenol, Estrogen-related receptor gamma | | Authors: | Yoon, H, Kim, J, Chin, J, Song, J, Cho, S.J. | | Deposit date: | 2019-08-07 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | An orally available inverse agonist of estrogen-related receptor gamma showed expanded efficacy for the radioiodine therapy of poorly differentiated thyroid cancer.
Eur.J.Med.Chem., 205, 2020
|
|
7S5B
 
 | |
3MYG
 
 | | Aurora A Kinase complexed with SCH 1473759 | | Descriptor: | 2-{ethyl[(5-{[6-methyl-3-(1H-pyrazol-4-yl)imidazo[1,2-a]pyrazin-8-yl]amino}isothiazol-3-yl)methyl]amino}-2-methylpropan-1-ol, Serine/threonine-protein kinase 6, TETRAETHYLENE GLYCOL | | Authors: | Hruza, A, Prosis, W, Ramanathan, L. | | Deposit date: | 2010-05-10 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Injectable Inhibitor of Aurora Kinases Based on the Imidazo-[1,2-a]-Pyrazine Core.
ACS Med Chem Lett, 1, 2010
|
|
2VUY
 
 | | Crystal structure of Glycogen Debranching exzyme TreX from Sulfolobus solfatarius | | Descriptor: | GLYCOGEN OPERON PROTEIN GLGX | | Authors: | Song, H.-N, Yoon, S.-M, Cha, H.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2008-06-02 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
4BVX
 
 | | Crystal structure of the AIMP3-MRS N-terminal domain complex with I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, EUKARYOTIC TRANSLATION ELONGATION FACTOR 1 EPSILON-1, METHIONINE--TRNA LIGASE, ... | | Authors: | Cho, H.Y, Seo, W.W, Cho, H.J, Kang, B.S. | | Deposit date: | 2013-06-29 | | Release date: | 2014-07-16 | | Last modified: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Assembly of Multi-tRNA Synthetase Complex Via Heterotetrameric Glutathione Transferase-Homology Domains.
J.Biol.Chem., 290, 2015
|
|
7VYT
 
 | | Crystal structure of human TIGIT(23-129) in complex with the scFv fragment of anti-TIGIT antibody MG1131 | | Descriptor: | CITRATE ANION, MG1131 heavy chain variable region, MG1131 light chain variable region, ... | | Authors: | Jeong, B.-S, Nam, H, Kim, M, Oh, B.-H. | | Deposit date: | 2021-11-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural and functional characterization of a monoclonal antibody blocking TIGIT.
Mabs, 14, 2022
|
|
5YC8
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS (Hg-derivative) | | Descriptor: | MERCURY (II) ION, Muscarinic acetylcholine receptor M2,Redesigned apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-06 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZK8
 
 | | Crystal structure of M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Redesigned apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZKB
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with AF-DX 384 | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-[2-[(2S)-2-[(dipropylamino)methyl]piperidin-1-yl]ethyl]-6-oxidanylidene-5H-pyrido[2,3-b][1,4]benzodiazepine-11-carboxamide | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
7N9W
 
 | |
7N8P
 
 | |
5ZKC
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZK3
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with QNB | | Descriptor: | (3R)-1-azabicyclo[2.2.2]oct-3-yl hydroxy(diphenyl)acetate, Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2 | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
7RP5
 
 | |
7RP6
 
 | |
7RO0
 
 | |
7RQ7
 
 | |
7RSB
 
 | |
7RNX
 
 | |
5I9J
 
 | | Structure of the cholesterol and lutein-binding domain of human STARD3 at 1.74A | | Descriptor: | 1,2-ETHANEDIOL, L(+)-TARTARIC ACID, SULFATE ION, ... | | Authors: | Horvath, M.P, Bernstein, P.S, Li, B, George, E.W, Tran, Q.T. | | Deposit date: | 2016-02-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of the lutein-binding domain of human StARD3 at 1.74 angstrom resolution and model of a complex with lutein.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
