2QHR
 
 | | Crystal structure of the 13F6-1-2 Fab fragment bound to its Ebola virus glycoprotein peptide epitope. | | 分子名称: | 13F6-1-2 Fab fragment V lambda x light chain, 13F6-1-2 Fab fragment heavy chain, Envelope glycoprotein peptide | | 著者 | Lee, J.E, Kuehne, A, Abelson, D.M, Fusco, M.L, Hart, M.K, Saphire, E.O. | | 登録日 | 2007-07-02 | | 公開日 | 2008-01-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Complex of a protective antibody with its Ebola virus GP peptide epitope: unusual features of a V lambda x light chain.
J.Mol.Biol., 375, 2008
|
|
3CSY
 
 | | Crystal structure of the trimeric prefusion Ebola virus glycoprotein in complex with a neutralizing antibody from a human survivor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein GP1, ... | | 著者 | Lee, J.E, Fusco, M.L, Hessell, A.J, Oswald, W.B, Burton, D.R, Saphire, E.O. | | 登録日 | 2008-04-10 | | 公開日 | 2008-07-08 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor.
Nature, 454, 2008
|
|
1Y6Q
 
 | | Cyrstal structure of MTA/AdoHcy nucleosidase complexed with MT-DADMe-ImmA | | 分子名称: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, CHLORIDE ION, MTA/SAH nucleosidase | | 著者 | Lee, J.E, Singh, V, Evans, G.B, Tyler, P.C, Furneaux, R.H, Cornell, K.A, Riscoe, M.K, Schramm, V.L, Howell, P.L. | | 登録日 | 2004-12-06 | | 公開日 | 2005-03-01 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
J.Biol.Chem., 280, 2005
|
|
1Y6R
 
 | | Crystal structure of MTA/AdoHcy nucleosidase complexed with MT-ImmA. | | 分子名称: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, MTA/SAH nucleosidase | | 著者 | Lee, J.E, Singh, V, Evans, G.B, Tyler, P.C, Furneaux, R.H, Cornell, K.A, Riscoe, M.K, Schramm, V.L, Howell, P.L. | | 登録日 | 2004-12-06 | | 公開日 | 2005-03-01 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
J.Biol.Chem., 280, 2005
|
|
1NC1
 
 | | Crystal structure of E. coli MTA/AdoHcy nucleosidase complexed with 5'-methylthiotubercidin (MTH) | | 分子名称: | 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, MTA/SAH nucleosidase | | 著者 | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | 登録日 | 2002-12-04 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of Escherichia coli 5'-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis.
J.Biol.Chem., 278, 2003
|
|
1NC3
 
 | | Crystal structure of E. coli MTA/AdoHcy nucleosidase complexed with formycin A (FMA) | | 分子名称: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, MTA/SAH nucleosidase | | 著者 | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | 登録日 | 2002-12-04 | | 公開日 | 2003-03-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of Escherichia coli 5'-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis.
J.Biol.Chem., 278, 2003
|
|
3INU
 
 | | Crystal structure of an unbound KZ52 neutralizing anti-Ebolavirus antibody. | | 分子名称: | GLYCEROL, KZ52 antibody fragment heavy chain, KZ52 antibody fragment light chain, ... | | 著者 | Lee, J.E, Fusco, M.L, Abelson, D.M, Hessell, A.J, Burton, D.R, Saphire, E.O. | | 登録日 | 2009-08-12 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Techniques and tactics used in determining the structure of the trimeric ebolavirus glycoprotein.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1JYS
 
 | | Crystal Structure of E. coli MTA/AdoHcy Nucleosidase | | 分子名称: | ADENINE, MTA/SAH nucleosidase | | 著者 | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | 登録日 | 2001-09-13 | | 公開日 | 2002-10-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of E. coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase reveals similarity to the purine nucleoside phosphorylases.
Structure, 9, 2001
|
|
1Z5N
 
 | | Crystal structure of MTA/AdoHcy nucleosidase Glu12Gln mutant complexed with 5-methylthioribose and adenine | | 分子名称: | 5-S-methyl-5-thio-alpha-D-ribofuranose, ADENINE, MTA/SAH nucleosidase | | 著者 | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | 登録日 | 2005-03-18 | | 公開日 | 2005-10-04 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
1Z5P
 
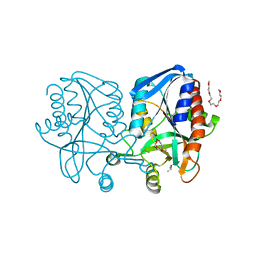 | | Crystal structure of MTA/AdoHcy nucleosidase with a ligand-free purine binding site | | 分子名称: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | 登録日 | 2005-03-18 | | 公開日 | 2005-10-04 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
1Z5O
 
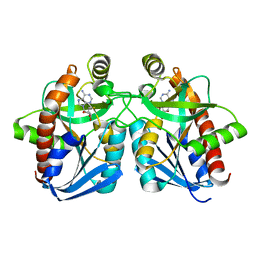 | | Crystal structure of MTA/AdoHcy nucleosidase Asp197Asn mutant complexed with 5'-methylthioadenosine | | 分子名称: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MTA/SAH nucleosidase | | 著者 | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | 登録日 | 2005-03-18 | | 公開日 | 2005-10-04 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
1SD1
 
 | | STRUCTURE OF HUMAN 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH FORMYCIN A | | 分子名称: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 5'-methylthioadenosine phosphorylase | | 著者 | Lee, J.E, Settembre, E.C, Cornell, K.A, Riscoe, M.K, Sufrin, J.R, Ealick, S.E, Howell, P.L. | | 登録日 | 2004-02-12 | | 公開日 | 2004-05-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structural Comparison of MTA Phosphorylase and MTA/AdoHcy Nucleosidase Explains Substrate Preferences and Identifies Regions Exploitable for Inhibitor Design.
Biochemistry, 43, 2004
|
|
1SD2
 
 | | STRUCTURE OF HUMAN 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH 5'-METHYLTHIOTUBERCIDIN | | 分子名称: | 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 5'-methylthioadenosine phosphorylase, SULFATE ION | | 著者 | Lee, J.E, Settembre, E.C, Cornell, K.A, Riscoe, M.K, Sufrin, J.R, Ealick, S.E, Howell, P.L. | | 登録日 | 2004-02-12 | | 公開日 | 2004-05-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Comparison of MTA Phosphorylase and MTA/AdoHcy Nucleosidase Explains Substrate Preferences and Identifies Regions Exploitable for Inhibitor Design.
Biochemistry, 43, 2004
|
|
2GMK
 
 | | Crystal structure of onconase double mutant with spontaneously-assembled (AMP) 4 stack | | 分子名称: | ADENOSINE MONOPHOSPHATE, P-30 protein | | 著者 | Bae, E, Lee, J.E, Raines, R.T, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-04-06 | | 公開日 | 2006-04-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis for catalysis by onconase.
J.Mol.Biol., 375, 2008
|
|
2I5S
 
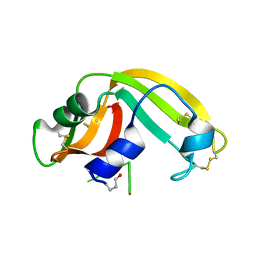 | | Crystal structure of onconase with bound nucleic acid | | 分子名称: | 5'-D(*A*(DU)P*GP*A)-3', P-30 protein | | 著者 | Bae, E, Lee, J.E, Raines, R.T, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-08-25 | | 公開日 | 2006-09-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for catalysis by onconase.
J.Mol.Biol., 375, 2008
|
|
3TIS
 
 | | Crystal structures of yrdA from Escherichia coli, a homologous protein of gamma-class carbonic anhydrases, show possible allosteric conformations | | 分子名称: | Protein YrdA, ZINC ION | | 著者 | Park, H.M, Chio, J.W, Lee, J.E, Jung, J.H, Kim, B.Y, Kim, J.S. | | 登録日 | 2011-08-21 | | 公開日 | 2012-08-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of the gamma-class carbonic anhydrase homologue YrdA suggest a possible allosteric switch
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TIO
 
 | | Crystal structures of yrdA from Escherichia coli, a homologous protein of gamma-class carbonic anhydrase, show possible allosteric conformations | | 分子名称: | PHOSPHATE ION, Protein YrdA, ZINC ION | | 著者 | Park, H.M, Choi, J.W, Lee, J.E, Jung, C.H, Kim, B.Y, Kim, J.S. | | 登録日 | 2011-08-21 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Structures of the gamma-class carbonic anhydrase homologue YrdA suggest a possible allosteric switch
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1AKP
 
 | | SEQUENTIAL 1H,13C AND 15N NMR ASSIGNMENTS AND SOLUTION CONFORMATION OF APOKEDARCIDIN | | 分子名称: | APOKEDARCIDIN | | 著者 | Constantine, K.L, Colson, K.L, Wittekind, M, Friedrichs, M.S, Zein, N, Tuttle, J, Langley, D.R, Leet, J.E, Schroeder, D.R, Lam, K.S, Farmer II, B.T, Metzler, W.J, Bruccoleri, R.E, Mueller, L. | | 登録日 | 1994-06-20 | | 公開日 | 1994-08-31 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Sequential 1H, 13C, and 15N NMR assignments and solution conformation of apokedarcidin.
Biochemistry, 33, 1994
|
|
7UJV
 
 | | Structure of PHD2 in complex with HIF2a-CODD | | 分子名称: | Egl nine homolog 1, Endothelial PAS domain-containing protein 1, FE (III) ION, ... | | 著者 | Ferens, F.G, Tarade, D, Lee, J.E, Ohh, M. | | 登録日 | 2022-03-31 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Deficiency in PHD2-mediated hydroxylation of HIF2 alpha underlies Pacak-Zhuang syndrome.
Commun Biol, 7, 2024
|
|
4J4J
 
 | |
4JF3
 
 | |
4JGS
 
 | |
5F4E
 
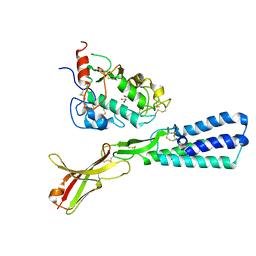 | | Crystal structure of the human sperm Izumo1 and egg Juno complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | 著者 | Aydin, H, Sultana, A, Lee, J.E. | | 登録日 | 2015-12-03 | | 公開日 | 2016-06-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular architecture of the human sperm IZUMO1 and egg JUNO fertilization complex.
Nature, 534, 2016
|
|
5F4V
 
 | |
5F4Q
 
 | | Crystal structure of the human egg surface protein Juno | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | 著者 | Aydin, H, Sultana, A, Lee, J.E. | | 登録日 | 2015-12-03 | | 公開日 | 2016-06-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular architecture of the human sperm IZUMO1 and egg JUNO fertilization complex.
Nature, 534, 2016
|
|
