6GLB
 
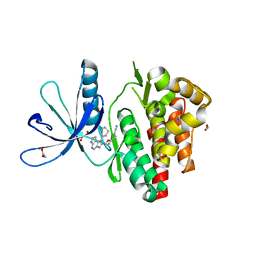 | | Crystal structure of JAK3 in complex with Compound 20 (FM484) | | Descriptor: | 1,2-ETHANEDIOL, 1-phenylurea, 3-[5-(3-cyclohexyl-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1(9),2(6),4,7,11-pentaen-4-yl)furan-2-yl]propanenitrile, ... | | Authors: | Chaikuad, A, Forster, M, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development, Optimization, and Structure-Activity Relationships of Covalent-Reversible JAK3 Inhibitors Based on a Tricyclic Imidazo[5,4- d]pyrrolo[2,3- b]pyridine Scaffold.
J. Med. Chem., 61, 2018
|
|
5LWM
 
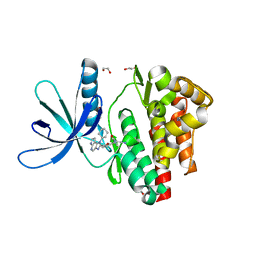 | | Crystal structure of JAK3 in complex with Compound 4 (FM381) | | Descriptor: | 1,2-ETHANEDIOL, 1-phenylurea, 2-cyano-3-[5-(3-cyclohexyl-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl)furan-2-yl]-~{N},~{N}-dimethyl-prop-2-enamide, ... | | Authors: | Chaikuad, A, Forster, M, Mukhopadhyay, S, Kupinska, K, Ellis, K, Mahajan, P, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-18 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selective JAK3 Inhibitors with a Covalent Reversible Binding Mode Targeting a New Induced Fit Binding Pocket.
Cell Chem Biol, 23, 2016
|
|
4X21
 
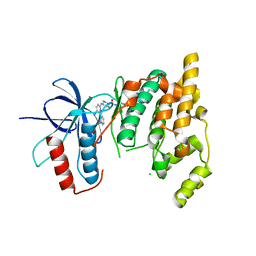 | | The MAP kinase JNK3 as target for halogen bonding | | Descriptor: | CHLORIDE ION, Mitogen-activated protein kinase 10, N-ethyl-4-{[4-(1H-indol-3-yl)-5-iodopyrimidin-2-yl]amino}piperidine-1-carboxamide | | Authors: | Lange, A, Buettner, F.M, Guenther, M.B, Zimmermann, M.O, Hennig, S, Laufer, S.A, Stehle, T, Boeckler, F. | | Deposit date: | 2014-11-25 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting the Gatekeeper MET146 of C-Jun N-Terminal Kinase 3 Induces a Bivalent Halogen/Chalcogen Bond.
J.Am.Chem.Soc., 137, 2015
|
|
9GV2
 
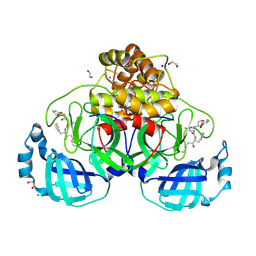 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalently bound inhibitor FP237 (compound 8p in publication) | | Descriptor: | (2S)-2-[2-(3-methoxyphenoxy)ethanoylamino]-4-methyl-N-[(2S)-3-oxidanylidene-1-phenyl-pentan-2-yl]pentanamide, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Strater, N, Sylvester, K, Muller, C.E, Flury, P, Kruger, N, Breidenbach, J, Guetschow, M, Laufer, S.A, Pillaiyar, T. | | Deposit date: | 2024-09-20 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Design, Synthesis, and Unprecedented Interactions of Covalent Dipeptide-Based Inhibitors of SARS-CoV-2 Main Protease and Its Variants Displaying Potent Antiviral Activity.
J.Med.Chem., 68, 2025
|
|
3ZYA
 
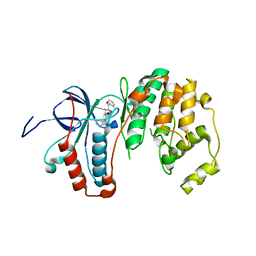 | | Human p38 MAP Kinase in Complex with 2-amino-phenylamino- dibenzosuberone | | Descriptor: | 2-AMINO-PHENYLAMINO-DIBENZOSUBERONE, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Romir, J, Koeberle, S.C, Laufer, S.A, Stehle, T. | | Deposit date: | 2011-08-18 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Skepinone-L is a Selective P38 Mitogen-Activated Protein Kinase Inhibitor.
Nat.Chem.Biol., 8, 2011
|
|
5LWN
 
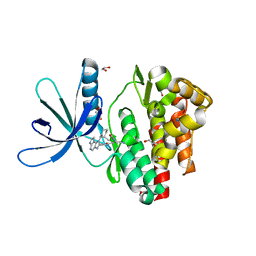 | | Crystal structure of JAK3 in complex with Compound 5 (FM409) | | Descriptor: | (2~{S})-2-cyano-~{N},~{N}-dimethyl-3-[5-[3-[(1~{S},2~{R})-2-methylcyclohexyl]-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl]furan-2-yl]propanamide, (~{Z})-2-cyano-~{N},~{N}-dimethyl-3-[5-[3-[(1~{S},2~{R})-2-methylcyclohexyl]-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl]furan-2-yl]prop-2-enamide, 1,2-ETHANEDIOL, ... | | Authors: | Chaikuad, A, Forster, M, Mukhopadhyay, S, Kupinska, K, Ellis, K, Mahajan, P, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-18 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective JAK3 Inhibitors with a Covalent Reversible Binding Mode Targeting a New Induced Fit Binding Pocket.
Cell Chem Biol, 23, 2016
|
|
6GL9
 
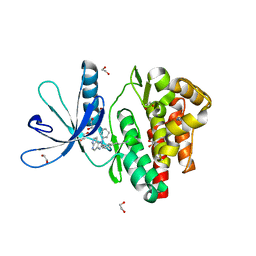 | | Crystal structure of JAK3 in complex with Compound 10 (FM475) | | Descriptor: | (~{E})-3-[3-(3-cyclohexyl-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1(9),2(6),4,7,11-pentaen-4-yl)phenyl]prop-2-enenitrile, 1,2-ETHANEDIOL, 1-phenylurea, ... | | Authors: | Chaikuad, A, Forster, M, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development, Optimization, and Structure-Activity Relationships of Covalent-Reversible JAK3 Inhibitors Based on a Tricyclic Imidazo[5,4- d]pyrrolo[2,3- b]pyridine Scaffold.
J. Med. Chem., 61, 2018
|
|
6GLA
 
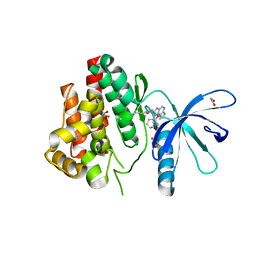 | | Crystal structure of JAK3 in complex with Compound 11 (FM481) | | Descriptor: | (~{E})-3-[5-(3-cyclohexyl-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1(9),2(6),4,7,11-pentaen-4-yl)furan-2-yl]prop-2-enenitrile, 1,2-ETHANEDIOL, 1-phenylurea, ... | | Authors: | Chaikuad, A, Forster, M, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Development, Optimization, and Structure-Activity Relationships of Covalent-Reversible JAK3 Inhibitors Based on a Tricyclic Imidazo[5,4- d]pyrrolo[2,3- b]pyridine Scaffold.
J. Med. Chem., 61, 2018
|
|
9F32
 
 | | Crystal structure of ULK1 with a covalent compound GCL 99 | | Descriptor: | N-[4-(1H-indazol-5-yl)phenyl]propanamide, Serine/threonine-protein kinase ULK1 | | Authors: | Wang, G.Q, Seidler, N, Gehringer, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-24 | | Release date: | 2025-01-08 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Protein Kinases' Cysteinome by Covalent Fragments.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9F81
 
 | | Crystal structure of RIOK2 with a covalent compound GCL 47 | | Descriptor: | N-(4-quinazolin-4-ylphenyl)propanamide, Serine/threonine-protein kinase RIO2 | | Authors: | Wang, G.Q, Wydra, V, Gehringer, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-05-06 | | Release date: | 2025-01-08 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Probing the Protein Kinases' Cysteinome by Covalent Fragments.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8P7J
 
 | | Crystal structure of MAP2K6 with a covalent compound GCL96 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 6, N-[3-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl]prop-2-enamide | | Authors: | Wang, G.Q, Seidler, N, Roehm, S, Gehringer, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-30 | | Release date: | 2023-07-05 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the Protein Kinases' Cysteinome by Covalent Fragments.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8PM3
 
 | | Crystal structure of MAP2K6 with a covalent compound GCL94 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 6, ~{N}-[3-(2-azanylpyridin-4-yl)phenyl]propanamide | | Authors: | Wang, G.Q, Seidler, N, Roehm, S, Gehringer, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-28 | | Release date: | 2023-09-20 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the Protein Kinases' Cysteinome by Covalent Fragments.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9F31
 
 | | Crystal structure of MELK with a covalent compound GCL 99 | | Descriptor: | Maternal embryonic leucine zipper kinase, N-[4-(1H-indazol-5-yl)phenyl]propanamide | | Authors: | Wang, G.Q, Seidler, N, Gehringer, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-24 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the Protein Kinases' Cysteinome by Covalent Fragments.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8FV4
 
 | | EGFR(T790M/V948R) in complex with compound 2 (LN5993) | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, N-{(3P)-3-[(4P)-4-(2-acetamidopyridin-4-yl)-2-(methylsulfanyl)-1H-imidazol-5-yl]phenyl}-11-oxo-10,11-dihydro-5H-dibenzo[b,e][1,4]diazepine-9-carboxamide, ... | | Authors: | Ogboo, B.C, Beyett, T.S, Eck, M.J, Heppner, D.E. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Linking ATP and allosteric sites to achieve superadditive binding with bivalent EGFR kinase inhibitors.
Commun Chem, 7, 2024
|
|
8FV3
 
 | | EGFR(T790M/V948R) in complex with compound 1 (LN4503) | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, N-{(4P)-4-[(4P)-5-{3-[(8-fluoro-11-oxo-5,11-dihydro-10H-dibenzo[b,e][1,4]diazepin-10-yl)methyl]phenyl}-2-(methylsulfanyl)-1H-imidazol-4-yl]pyridin-2-yl}acetamide, ... | | Authors: | Ogboo, B.C, Heppner, D.E. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Linking ATP and allosteric sites to achieve superadditive binding with bivalent EGFR kinase inhibitors.
Commun Chem, 7, 2024
|
|
8HI9
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with Robinetin | | Descriptor: | 3,7-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-11-19 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of Polyphenolic Natural Products as SARS-CoV-2 M pro Inhibitors for COVID-19.
Pharmaceuticals, 16, 2023
|
|
9HHW
 
 | | Crystal structure of TTBK1 with a covalent compound GCL95 | | Descriptor: | Tau-tubulin kinase 1, ~{N}-[2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yl)phenyl]propanamide | | Authors: | Wang, G, Seidler, N, Gehringer, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-22 | | Release date: | 2025-01-08 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the Protein Kinases' Cysteinome by Covalent Fragments.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
3UVP
 
 | | Human p38 MAP Kinase in Complex with a Benzamide Substituted Benzosuberone | | Descriptor: | Mitogen-activated protein kinase 14, N-{2-fluoro-5-[(5-oxo-6,7,8,9-tetrahydro-5H-benzo[7]annulen-2-yl)amino]phenyl}benzamide, octyl beta-D-glucopyranoside | | Authors: | Mayer-Wrangowski, S.C, Richters, A, Gruetter, C, Rauh, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Hinge Glycine Flip and the Activation Loop: Novel Approach to Potent p38 alpha Inhibitors.
J.Med.Chem., 55, 2012
|
|
3UVQ
 
 | | Human p38 MAP Kinase in Complex with a Dibenzosuberone Derivative | | Descriptor: | Mitogen-activated protein kinase 14, N-{5-[(7-{[(2R)-2,3-dihydroxypropyl]oxy}-5-oxo-10,11-dihydro-5H-dibenzo[a,d][7]annulen-2-yl)amino]-2-fluorophenyl}benzamide, octyl beta-D-glucopyranoside | | Authors: | Mayer-Wrangowski, S.C, Richters, A, Gruetter, C, Rauh, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dibenzosuberones as p38 mitogen-activated protein kinase inhibitors with low ATP competitiveness and outstanding whole blood activity.
J.Med.Chem., 56, 2013
|
|
7UKV
 
 | | Wild type EGFR in complex with Lazertinib (YH25448) | | Descriptor: | Epidermal growth factor receptor, N-[5-{[(4P)-4-{4-[(dimethylamino)methyl]-3-phenyl-1H-pyrazol-1-yl}pyrimidin-2-yl]amino}-4-methoxy-2-(morpholin-4-yl)phenyl]propanamide | | Authors: | Beyett, T.S, Pham, C, Eck, M.J, Heppner, D.E. | | Deposit date: | 2022-04-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Inhibition of Mutant EGFR with Lazertinib (YH25448).
Acs Med.Chem.Lett., 13, 2022
|
|
7UKW
 
 | | EGFR(T790M/V948R) in complex with Lazertinib (YH25448) | | Descriptor: | Epidermal growth factor receptor, N-[5-{[(4P)-4-{4-[(dimethylamino)methyl]-3-phenyl-1H-pyrazol-1-yl}pyrimidin-2-yl]amino}-4-methoxy-2-(morpholin-4-yl)phenyl]propanamide | | Authors: | Pham, C.D, Heppner, D.E. | | Deposit date: | 2022-04-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Inhibition of Mutant EGFR with Lazertinib (YH25448).
Acs Med.Chem.Lett., 13, 2022
|
|
7X6K
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3w | | Descriptor: | 1H-indole-2-carbaldehyde, 3C-like proteinase | | Authors: | Su, H, Nie, T, Xie, H, Li, Z.W, Li, M.J, Xu, Y. | | Deposit date: | 2022-03-07 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Small-Molecule Thioesters as SARS-CoV-2 Main Protease Inhibitors: Enzyme Inhibition, Structure-Activity Relationships, Antiviral Activity, and X-ray Structure Determination.
J.Med.Chem., 65, 2022
|
|
7X6J
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3af | | Descriptor: | 3C-like proteinase, quinoline-2-carboxylic acid | | Authors: | Su, H, Nie, T, Xie, H, Li, Z.W, Li, M.J, Xu, Y. | | Deposit date: | 2022-03-07 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small-Molecule Thioesters as SARS-CoV-2 Main Protease Inhibitors: Enzyme Inhibition, Structure-Activity Relationships, Antiviral Activity, and X-ray Structure Determination.
J.Med.Chem., 65, 2022
|
|
6WXN
 
 | | EGFR(T790M/V948R) in complex with LN3844 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Design of a "Two-in-One" Mutant-Selective Epidermal Growth Factor Receptor Inhibitor That Spans the Orthosteric and Allosteric Sites.
J.Med.Chem., 65, 2022
|
|
4L8M
 
 | | Human p38 MAP kinase in complex with a Dibenzoxepinone | | Descriptor: | Mitogen-activated protein kinase 14, N-[2-fluoro-5-({9-[2-(morpholin-4-yl)ethoxy]-11-oxo-6,11-dihydrodibenzo[b,e]oxepin-3-yl}amino)phenyl]benzamide, octyl beta-D-glucopyranoside | | Authors: | Richters, A, Mayer-Wrangowski, S.C, Gruetter, C, Rauh, D. | | Deposit date: | 2013-06-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metabolically Stable Dibenzo[b,e]oxepin-11(6H)-ones as Highly Selective p38 MAP Kinase Inhibitors: Optimizing Anti-Cytokine Activity in Human Whole Blood.
J.Med.Chem., 56, 2013
|
|
