6YCA
 
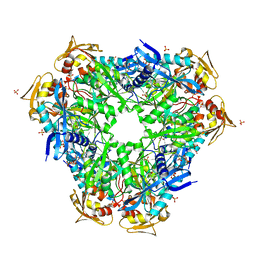 | | Crystal structure of Eis1 from Mycobacterium abscessus | | Descriptor: | ACETYL COENZYME *A, SULFATE ION, Uncharacterized N-acetyltransferase D2E76_00625 | | Authors: | Blaise, M, Ung, K.L. | | Deposit date: | 2020-03-18 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of the N-acetyltransferase Eis1 from Mycobacterium abscessus reveals the molecular determinants of its incapacity to modify aminoglycosides.
Proteins, 89, 2021
|
|
7N11
 
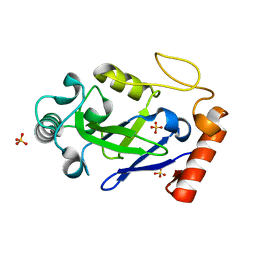 | |
7N12
 
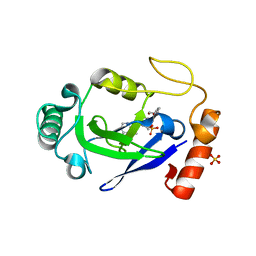 | | Crystal structure of the M. abscessus LeuRS editing domain in complex with epetraborole-AMP adduct | | Descriptor: | Leucine--tRNA ligase, SULFATE ION, [(1S,5R,6R,7'S,8R)-7'-(aminomethyl)-6-(6-aminopurin-9-yl)-2'-(3-oxidanylpropoxy)spiro[2,4,7-trioxa-3-boranuidabicyclo[3.3.0]octane-3,9'-8-oxa-9-boranuidabicyclo[4.3.0]nona-1(6),2,4-triene]-8-yl]methyl dihydrogen phosphate | | Authors: | Kalthoff, E, Schmeing, M. | | Deposit date: | 2021-05-26 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Efficacy of epetraborole against Mycobacterium abscessus is increased with norvaline.
Plos Pathog., 17, 2021
|
|
7Q3A
 
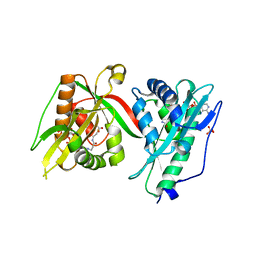 | | Crystal structure of MAB_4324 a tandem repeat GNAT from Mycobacterium abscessus | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Blaise, M, Alsarraf, M.A.B. | | Deposit date: | 2021-10-27 | | Release date: | 2022-05-04 | | Last modified: | 2022-07-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical, structural, and functional studies reveal that MAB_4324c from Mycobacterium abscessus is an active tandem repeat N-acetyltransferase.
Febs Lett., 596, 2022
|
|
6T84
 
 | |
3F4B
 
 | |
5T2U
 
 | | short chain dehydrogenase/reductase family protein | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, short chain dehydrogenase/reductase family protein | | Authors: | Mickael, B, Van Wyk, N, Baneres-Roquet, F, Yann, G, Laurent, K. | | Deposit date: | 2016-08-24 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of NADP(+) triggers an open-to-closed transition in a mycobacterial FabG beta-ketoacyl-ACP reductase.
Biochem. J., 474, 2017
|
|
7U0O
 
 | |
7U0M
 
 | |
7AGO
 
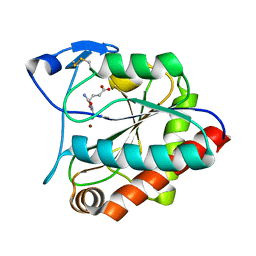 | | crystal structure of the N-acetylmuramyl-L-alanine amidase, Ami1, from Mycobacterium abscessus bound to L-Alanine-D-isoglutamine | | Descriptor: | ALANINE, D-alpha-glutamine, N-acetylmuramoyl-L-alanine amidase, ... | | Authors: | Blaise, M. | | Deposit date: | 2020-09-23 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional Characterization of the N -Acetylmuramyl-l-Alanine Amidase, Ami1, from Mycobacterium abscessus .
Cells, 9, 2020
|
|
7AGM
 
 | | Crystal structure of the N-acetylmuramyl-L-alanine amidase, Ami1, from Mycobacterium smegmatis | | Descriptor: | N-acetylmuramoyl-L-alanine amidase, ZINC ION | | Authors: | Blaise, M, Alsarraf, M.A.B. | | Deposit date: | 2020-09-23 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Functional Characterization of the N -Acetylmuramyl-l-Alanine Amidase, Ami1, from Mycobacterium abscessus .
Cells, 9, 2020
|
|
7AGL
 
 | |
6SHW
 
 | | N-terminal domain of Drosophila X Virus VP3 | | Descriptor: | SULFATE ION, Structural polyprotein | | Authors: | Ferrero, D.S, Garriga, D, Guerra, P, Uson, I, Verdaguer, N. | | Deposit date: | 2019-08-08 | | Release date: | 2020-11-18 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and dsRNA-binding activity of the Birnavirus Drosophila X Virus VP3 protein.
J.Virol., 2020
|
|
6SI6
 
 | | N-terminal domain of Drosophila X virus VP3 | | Descriptor: | GLYCEROL, IMIDAZOLE, Structural polyprotein | | Authors: | Ferrero, D.S, Garriga, D, Guerra, P, Uson, I, Verdaguer, N. | | Deposit date: | 2019-08-08 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and dsRNA-binding activity of the Birnavirus Drosophila X Virus VP3 protein.
J.Virol., 2020
|
|
7L6C
 
 | |
7K73
 
 | |
7KLI
 
 | |
2NV6
 
 | |
2GP6
 
 | |
2KB3
 
 | | NMR Structure of the phosphorylated form of OdhI, pOdhI. | | Descriptor: | Oxoglutarate dehydrogenase inhibitor | | Authors: | Barthe, P, Roumestand, C, Canova, M, Hurard, C, Molle, V, Cohen-Gonsaud, M. | | Deposit date: | 2008-11-20 | | Release date: | 2009-05-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Dynamic and Structural Characterization of a Bacterial FHA Protein Reveals a New Autoinhibition Mechanism.
Structure, 17, 2009
|
|
2KFS
 
 | | NMR structure of Rv2175c | | Descriptor: | Conserved hypothetical regulatory protein | | Authors: | Barthe, P, Cohen-Gonsaud, M, Roumestand, C, Molle, V. | | Deposit date: | 2009-02-27 | | Release date: | 2009-05-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Mycobacterium tuberculosis Ser/Thr Kinase Substrate Rv2175c Is a DNA-binding Protein Regulated by Phosphorylation.
J.Biol.Chem., 284, 2009
|
|
2KB4
 
 | | NMR structure of the unphosphorylated form of OdhI, OdhI. | | Descriptor: | Oxoglutarate dehydrogenase inhibitor | | Authors: | Barthe, P, Roumestand, C, Canova, M, Hurard, C, Molle, V, Cohen-Gonsaud, M. | | Deposit date: | 2008-11-20 | | Release date: | 2009-05-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Dynamic and Structural Characterization of a Bacterial FHA Protein Reveals a New Autoinhibition Mechanism.
Structure, 17, 2009
|
|
2GEJ
 
 | | Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP-Man | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, PHOSPHATIDYLINOSITOL MANNOSYLTRANSFERASE (PimA) | | Authors: | Guerin, M.E, Buschiazzo, A, Kordulakova, J, Jackson, M, Alzari, P.M. | | Deposit date: | 2006-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria.
J.Biol.Chem., 282, 2007
|
|
2GEK
 
 | | Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHATIDYLINOSITOL MANNOSYLTRANSFERASE (PimA) | | Authors: | Guerin, M.E, Buschiazzo, A, Kordulakova, J, Jackson, M, Alzari, P.M. | | Deposit date: | 2006-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria.
J.Biol.Chem., 282, 2007
|
|
