8FNU
 
 | | Structure of RdrA from Streptococcus suis RADAR defense system | | Descriptor: | KAP NTPase domain-containing protein | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FNT
 
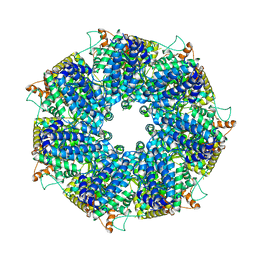 | | Structure of RdrA from Escherichia coli RADAR defense system | | Descriptor: | Archaeal ATPase | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FNV
 
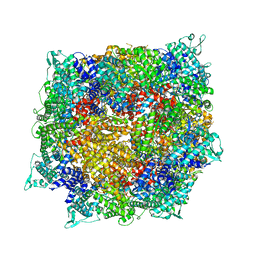 | | Structure of RdrB from Escherichia coli RADAR defense system | | Descriptor: | Adenosine deaminase, ZINC ION | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8GJX
 
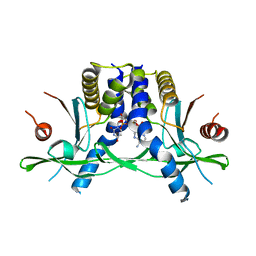 | | Structure of the human STING receptor bound to 2'3'-cUA | | Descriptor: | 2'3'-cUA, Stimulator of interferon genes protein | | Authors: | Morehouse, B.R, Li, Y, Slavik, K.M, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJZ
 
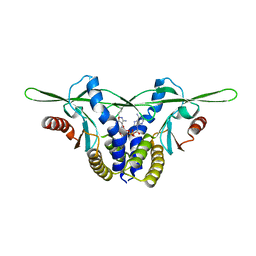 | | Structure of a STING receptor from S. pistillata Sp-STING1 bound to 2'3'-cUA | | Descriptor: | 2'3'-cUA, Stimulator of interferon genes protein | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJY
 
 | | Structure of a cGAS-like receptor Sp-cGLR1 from S. pistillata | | Descriptor: | cGAS-like receptor 1 | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJW
 
 | | Structure of a cGAS-like receptor Cv-cGLR1 from C. virginica | | Descriptor: | SULFATE ION, cGAS-like receptor 1 | | Authors: | Li, Y, Morehouse, B.R, Slavik, K.M, Liu, J, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8SL0
 
 | |
7N51
 
 | |
7N50
 
 | |
7N52
 
 | |
8GBE
 
 | |
8SME
 
 | | Structure of SPO1 phage Tad2 in apo state | | Descriptor: | Gp34.65 | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SMG
 
 | | Structure of SPO1 phage Tad2 in complex with 1''-2' gcADPR | | Descriptor: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, Gp34.65 | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SMF
 
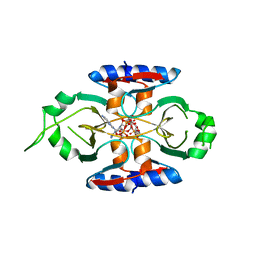 | | Structure of SPO1 phage Tad2 in complex with 1''-3' gcADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, Gp34.65, MAGNESIUM ION | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SMD
 
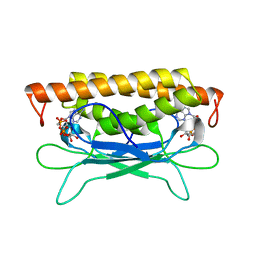 | | Structure of Clostridium botulinum prophage Tad1 in complex with 1''-3' gcADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, ABC transporter ATPase | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SM3
 
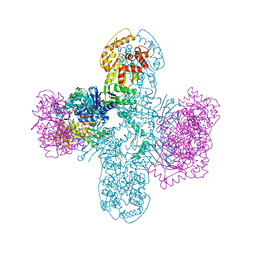 | | Structure of Bacillus cereus VD045 Gabija GajA-GajB Complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB, SULFATE ION | | Authors: | Antine, S.P, Mooney, S.E, Johnson, A.G, Kranzusch, P.J. | | Deposit date: | 2023-04-25 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of Gabija anti-phage defence and viral immune evasion.
Nature, 625, 2024
|
|
8T9N
 
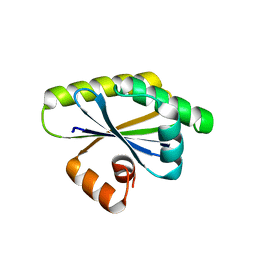 | | Bacillus subtilis RsgI GGG mutant | | Descriptor: | Anti-sigma-I factor RsgI | | Authors: | Brogan, A.P, Habib, C, Hobbs, S.J, Kranzusch, P.J, Rudner, D.Z. | | Deposit date: | 2023-06-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacterial SEAL domains undergo autoproteolysis and function in regulated intramembrane proteolysis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5UQD
 
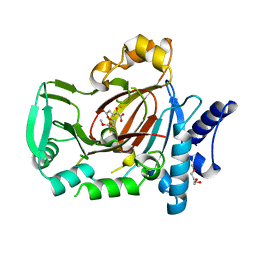 | | DPY-21 in complex with Fe(II) and alpha-Ketoglutarate | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-OXOGLUTARIC ACID, DumPY: shorter than wild-type, ... | | Authors: | Brejc, K, Bian, Q, Uzawa, S, Wheeler, B.S, Anderson, E.C, King, D.S, Kranzusch, P.J, Preston, C.G, Meyer, B.J. | | Deposit date: | 2017-02-07 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Dynamic Control of X Chromosome Conformation and Repression by a Histone H4K20 Demethylase.
Cell, 171, 2017
|
|
5VGB
 
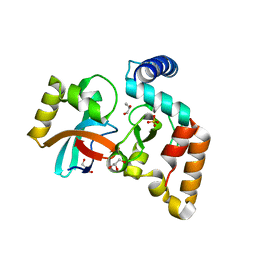 | | Crystal structure of NmeCas9 HNH domain bound to anti-CRISPR AcrIIC1 | | Descriptor: | Anti-CRISPR protein (AcrIIC1), CRISPR-associated endonuclease Cas9, GLYCEROL, ... | | Authors: | Harrington, L.B, Doxzen, K.W, Ma, E, Knott, G.J, Kranzusch, P.J, Doudna, J.A. | | Deposit date: | 2017-04-10 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | A Broad-Spectrum Inhibitor of CRISPR-Cas9.
Cell, 170, 2017
|
|
8SRZ
 
 | |
8SS1
 
 | |
7R65
 
 | |
7TQO
 
 | |
7TQQ
 
 | | Structure of human TREX1-DNA complex | | Descriptor: | DNA (5'-D(P*TP*TP*GP*GP*CP*CP*GP*GP*CP*CP*AP*TP*C)-3'), Three-prime repair exonuclease 1 | | Authors: | Zhou, W, Richmond-Buccola, D, Kranzusch, P.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of human TREX1 DNA degradation and autoimmune disease.
Nat Commun, 13, 2022
|
|
