4X1J
 
 | | X-ray crystal structure of the dimeric BMP antagonist NBL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroblastoma suppressor of tumorigenicity 1 | | Authors: | Thompson, T.B, Nolan, K, Kattamuri, C. | | Deposit date: | 2014-11-24 | | Release date: | 2015-01-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Neuroblastoma Suppressor of Tumorigenicity 1 (NBL1): INSIGHTS FOR THE FUNCTIONAL VARIABILITY ACROSS BONE MORPHOGENETIC PROTEIN (BMP) ANTAGONISTS.
J.Biol.Chem., 290, 2015
|
|
7MRZ
 
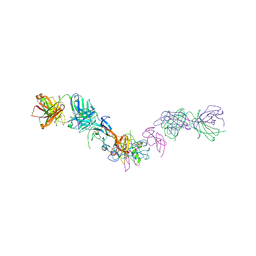 | | Structure of GDF11 bound to fused ActRIIB-ECD and Alk4-ECD with Anti-ActRIIB Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B,Activin receptor type-1B, Fab Heavy Chain, ... | | Authors: | Goebel, E.J, Kattamuri, C, Gipson, G.R, Thompson, T.B. | | Deposit date: | 2021-05-10 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of activin ligand traps using natural sets of type I and type II TGF beta receptors.
Iscience, 25, 2022
|
|
4JPH
 
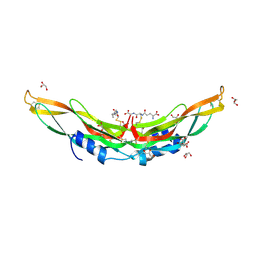 | | Crystal structure of Protein Related to DAN and Cerberus (PRDC) | | Descriptor: | CITRIC ACID, GLUTATHIONE, GLYCEROL, ... | | Authors: | Deng, X, Nolan, K.T, Kattamuri, C, Thompson, T.B. | | Deposit date: | 2013-03-19 | | Release date: | 2013-07-24 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of protein related to dan and cerberus: insights into the mechanism of bone morphogenetic protein antagonism.
Structure, 21, 2013
|
|
7OLY
 
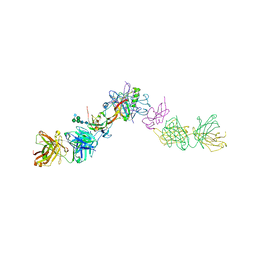 | | Structure of activin A in complex with an ActRIIB-Alk4 fusion reveal insight into activin receptor interactions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-1B, ... | | Authors: | Hakansson, M, Rose, N.C, Castonguay, R, Logan, D.T, Krishnan, L. | | Deposit date: | 2021-05-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.265 Å) | | Cite: | Structures of activin ligand traps using natural sets of type I and type II TGF beta receptors.
Iscience, 25, 2022
|
|
8E3G
 
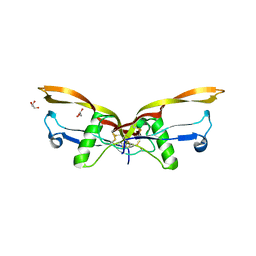 | | BMP2/GDF5 heterodimer | | Descriptor: | Bone morphogenetic protein 2, GLYCEROL, Growth/differentiation factor 5 | | Authors: | Gipson, G.R, Nolan, K.T, Thompson, T.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Formation and characterization of BMP2/GDF5 and BMP4/GDF5 heterodimers.
Bmc Biol., 21, 2023
|
|
5HK5
 
 | |
3B4V
 
 | | X-Ray structure of Activin in complex with FSTL3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Inhibin beta A chain, ... | | Authors: | Thompson, T.B. | | Deposit date: | 2007-10-24 | | Release date: | 2008-09-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The structure of FSTL3.activin A complex. Differential binding of N-terminal domains influences follistatin-type antagonist specificity.
J.Biol.Chem., 283, 2008
|
|
3SEK
 
 | | Crystal Structure of the Myostatin:Follistatin-like 3 Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Follistatin-related protein 3, Growth/differentiation factor 8 | | Authors: | Cash, J.N, Thompson, T.B. | | Deposit date: | 2011-06-10 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure of myostatinfollistatin-like 3: N-terminal domains of follistatin-type molecules exhibit alternate modes of binding.
J.Biol.Chem., 287, 2012
|
|
