2KXX
 
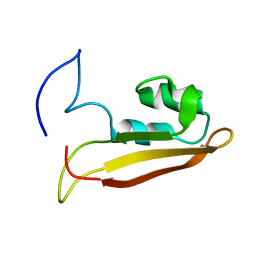 | | NMR Structure of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex | | Descriptor: | Small protein A | | Authors: | Kim, K, Okon, M, Escobar, E, Kang, H, McIntosh, L, Paetzel, M. | | Deposit date: | 2010-05-13 | | Release date: | 2011-01-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex.
Biochemistry, 50, 2011
|
|
2LZY
 
 | |
2LZX
 
 | |
7YOT
 
 | |
7YOU
 
 | |
7YOV
 
 | |
3ID4
 
 | | Crystal Structure of RseP PDZ2 domain fused GKASPV peptide | | Descriptor: | Regulator of sigma E protease | | Authors: | Li, X, Wang, B, Feng, L, Wang, J, Shi, Y. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ID2
 
 | | Crystal Structure of RseP PDZ2 domain | | Descriptor: | IODIDE ION, Regulator of sigma E protease | | Authors: | Li, X, Wang, B, Feng, L, Wang, J, Shi, Y. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ID1
 
 | | Crystal Structure of RseP PDZ1 domain | | Descriptor: | Regulator of sigma E protease | | Authors: | Li, X, Wang, B, Feng, L, Wang, J, Shi, Y. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ID3
 
 | | Crystal Structure of RseP PDZ2 I304A domain | | Descriptor: | Regulator of sigma E protease | | Authors: | Li, X, Wang, B, Feng, L, Wang, J, Shi, Y. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8HTI
 
 | | Human Consensus Olfactory Receptor OR52c in Complex with Octanoic Acid (OCA) and G Protein | | Descriptor: | Consensus Olfactory Receptor OR52c, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Choi, C.W, Bae, J, Choi, H.-J, Kim, J. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
8J46
 
 | | Human Consensus Olfactory Receptor OR52c in apo state, OR52c-bRIL | | Descriptor: | Olfactory receptor OR52c,Soluble cytochrome b562 | | Authors: | Choi, C.W, Bae, J, Choi, H.-J, Kim, J. | | Deposit date: | 2023-04-19 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
2GIR
 
 | | Hepatitis C virus RNA-dependent RNA polymerase NS5B with NNI-1 inhibitor | | Descriptor: | 3-{ISOPROPYL[(TRANS-4-METHYLCYCLOHEXYL)CARBONYL]AMINO}-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA-directed RNA polymerase | | Authors: | Harris, S.F. | | Deposit date: | 2006-03-29 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus.
J.Virol., 80, 2006
|
|
2GIQ
 
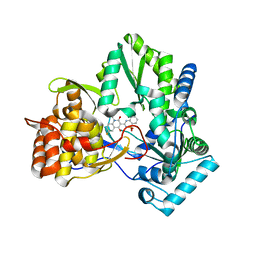 | | Hepatitis C virus RNA-dependent RNA polymerase NS5B with NNI-2 inhibitor | | Descriptor: | 1-(2-CYCLOPROPYLETHYL)-3-(1,1-DIOXIDO-2H-1,2,4-BENZOTHIADIAZIN-3-YL)-6-FLUORO-4-HYDROXYQUINOLIN-2(1H)-ONE, RNA-directed RNA polymerase | | Authors: | Harris, S.F. | | Deposit date: | 2006-03-29 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus.
J.Virol., 80, 2006
|
|
5GLG
 
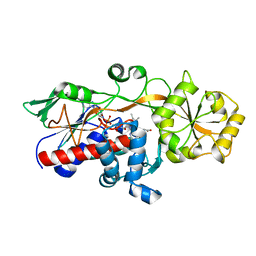 | |
5ZYN
 
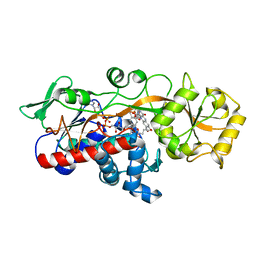 | | Fumarate reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fumarate reductase 2, ... | | Authors: | Park, H.H, Kim, C.M. | | Deposit date: | 2018-05-25 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular basis of maintaining an oxidizing environment under anaerobiosis by soluble fumarate reductase.
Nat Commun, 9, 2018
|
|
8E94
 
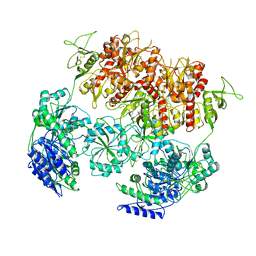 | | PYD-106-bound Human GluN1a-GluN2C NMDA receptor in intact conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E93
 
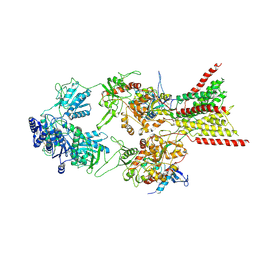 | | D-cycloserine and glutamate bound Human GluN1a-GluN2C NMDA receptor in splayed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E97
 
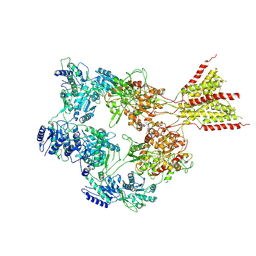 | | PYD-106-bound Human GluN1a-GluN2C NMDA receptor in splayed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E99
 
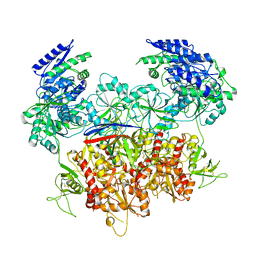 | | Human GluN1a-GluN2A-GluN2C triheteromeric NMDA receptor in complex with Nb-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E92
 
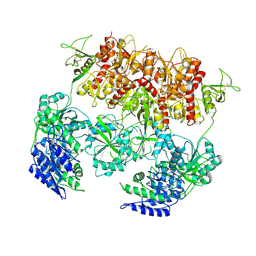 | | D-cycloserine and glutamate bound Human GluN1a-GluN2C NMDA receptor in intact conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E98
 
 | | D-cycloserine and glutamate bound Human GluN1a-GluN2C NMDA receptor in nanodisc - intact conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
7JX4
 
 | | Crystal Structure of N-Lysine Peptoid-modified Collagen Triple Helix | | Descriptor: | Collagen mimetic peptide with N-Lysine guest | | Authors: | Yu, M.S, Whitby, F.G, Hill, C.P, Kessler, J.L, Yang, L.D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Peptoid Residues Make Diverse, Hyperstable Collagen Triple-Helices.
J.Am.Chem.Soc., 143, 2021
|
|
7JX5
 
 | | Crystal Structure of N-Phenylalanine Peptoid-modified Collagen Triple Helix | | Descriptor: | Collagen mimetic peptide with N-Phenylalanine guest | | Authors: | Yu, M.S, Whitby, F.G, Hill, C.P, Kessler, J.L, Yang, L.D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peptoid Residues Make Diverse, Hyperstable Collagen Triple-Helices
J.Am.Chem.Soc., 143, 2021
|
|
8FN0
 
 | | CryoEM structure of Go-coupled NTSR1 with a biased allosteric modulator | | Descriptor: | 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Krumm, B.E, DiBerto, J.F, Olsen, R.H.J, Kang, H, Slocum, S.T, Zhang, S, Strachan, R.T, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-12-26 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Neurotensin Receptor Allosterism Revealed in Complex with a Biased Allosteric Modulator.
Biochemistry, 62, 2023
|
|
