6C5N
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase with hydroxyoxamate inhibitor 1 | | Descriptor: | (cyclopentylamino)(oxo)acetic acid, IMIDAZOLE, Ketol-acid reductoisomerase (NADP(+)), ... | | Authors: | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schembri, M.A, McGeary, R.P. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductosiomerase (KARI) inhibitor
To Be Published
|
|
6BUL
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase with hydroxyoxamate inhibitor 2 | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schembri, M.A, McFeary, R.P. | | Deposit date: | 2017-12-10 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductoisomerase (KARI) inhibitors
To Be Published
|
|
6C55
 
 | | Crystal structure of Staphylococcus aureus Ketol-acid Reductosimerrase with hydroxyoxamate inhibitor 3 | | Descriptor: | (cyclohexylamino)(oxo)acetic acid, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, ... | | Authors: | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schmbri, M, McGeary, R.P. | | Deposit date: | 2018-01-14 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductoisomerase (KARI) inhibitors
To Be Published
|
|
8OO2
 
 | | ChdA complex with amido-chelocardin | | Descriptor: | 2-carboxamido-2-deacetyl-chelocardin, MAGNESIUM ION, Putative transcriptional regulator | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2023-04-04 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Revision of the Absolute Configurations of Chelocardin and Amidochelocardin.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6ZYL
 
 | | non-heme monooxygenase; ThoJ apo | | Descriptor: | L(+)-TARTARIC ACID, Uncharacterized protein | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2020-08-02 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Non-Heme Monooxygenase ThoJ Catalyzes Thioholgamide beta-Hydroxylation.
Acs Chem.Biol., 15, 2020
|
|
6ZYK
 
 | | Non-heme monooxygenase, ThoJ-Ni complex | | Descriptor: | L(+)-TARTARIC ACID, Monooxygenase, NICKEL (II) ION | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2020-08-02 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Non-Heme Monooxygenase ThoJ Catalyzes Thioholgamide beta-Hydroxylation.
Acs Chem.Biol., 15, 2020
|
|
6NSQ
 
 | | Crystal structure of BRAF kinase domain bound to the inhibitor 2l | | Descriptor: | 5-[(4-amino-1-ethyl-1H-pyrazolo[3,4-d]pyrimidin-3-yl)ethynyl]-N-(4-chlorophenyl)-6-methylisoquinolin-1-amine, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Kurinov, I, Assadieskandar, A, Yu, C, Zhang, C, Sicheri, F. | | Deposit date: | 2019-01-25 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rigidification Dramatically Improves Inhibitor Selectivity for RAF Kinases.
Acs Med.Chem.Lett., 10, 2019
|
|
6T6H
 
 | | Apo structure of the Bottromycin epimerase BotH | | Descriptor: | BotH, SODIUM ION, SULFATE ION | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2019-10-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | The bottromycin epimerase BotH defines a group of atypical alpha / beta-hydrolase-fold enzymes.
Nat.Chem.Biol., 16, 2020
|
|
6T6Z
 
 | |
6T6Y
 
 | |
6T70
 
 | |
6T6X
 
 | | Structure of the Bottromycin epimerase BotH in complex with substrate | | Descriptor: | (4~{R})-2-[(1~{R})-1-[[(2~{S})-2-[[(2~{S})-3-methyl-2-[[(4~{Z},6~{S},9~{S},12~{S})-2,8,11-tris(oxidanylidene)-6,9-di(propan-2-yl)-1,4,7,10-tetrazabicyclo[10.3.0]pentadec-4-en-5-yl]amino]butanoyl]amino]-3-phenyl-propanoyl]amino]-3-oxidanyl-3-oxidanylidene-propyl]-4,5-dihydro-1,3-thiazole-4-carboxylic acid, BotH | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2019-10-20 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The bottromycin epimerase BotH defines a group of atypical alpha / beta-hydrolase-fold enzymes.
Nat.Chem.Biol., 16, 2020
|
|
6I5S
 
 | | AH, Bottromycin amidohydrolase | | Descriptor: | CHLORIDE ION, GLYCEROL, ZINC ION, ... | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2018-11-14 | | Release date: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Thiazoline-Specific Amidohydrolase PurAH Is the Gatekeeper of Bottromycin Biosynthesis.
J.Am.Chem.Soc., 141, 2019
|
|
6H96
 
 | | AlbA-albicidin complex, albicidin resistance protein | | Descriptor: | 4-[[4-[[4-[(3~{S})-5-azanyl-3-[[4-[[(~{E})-3-(4-hydroxyphenyl)-2-methyl-prop-2-enoyl]amino]phenyl]carbonylamino]-2-oxidanylidene-3~{H}-pyrrol-1-yl]phenyl]carbonylamino]-3-methoxy-2-oxidanyl-phenyl]carbonylamino]-3-methoxy-2-oxidanyl-benzoic acid, Albicidin resistance protein, SULFATE ION | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2018-08-03 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Adaptation of a Bacterial Multidrug Resistance System Revealed by the Structure and Function of AlbA.
J.Am.Chem.Soc., 140, 2018
|
|
6H95
 
 | | AlbA, albicidin resistance protein | | Descriptor: | Albicidin resistance protein | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2018-08-03 | | Release date: | 2018-11-21 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adaptation of a Bacterial Multidrug Resistance System Revealed by the Structure and Function of AlbA.
J.Am.Chem.Soc., 140, 2018
|
|
6HAI
 
 | | AlbAM131A mutant in complex with albicidin , albicidin resistance protein | | Descriptor: | 4-[[4-[[4-[(3~{S})-3-[[4-[[(~{E})-3-(4-hydroxyphenyl)-2-methyl-prop-2-enoyl]amino]phenyl]carbonylamino]-2,5-bis(oxidanylidene)pyrrolidin-1-yl]phenyl]carbonylamino]-3-methoxy-2-oxidanyl-phenyl]carbonylamino]-3-methoxy-2-oxidanyl-benzoic acid, Albicidin resistance protein, SULFATE ION | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2018-08-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Adaptation of a Bacterial Multidrug Resistance System Revealed by the Structure and Function of AlbA.
J.Am.Chem.Soc., 140, 2018
|
|
6H97
 
 | | AlbAT99V mutant , albicidin resistance protein | | Descriptor: | Albicidin resistance protein | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2018-08-03 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Adaptation of a Bacterial Multidrug Resistance System Revealed by the Structure and Function of AlbA.
J.Am.Chem.Soc., 140, 2018
|
|
6CAD
 
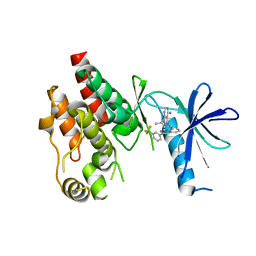 | | Crystal structure of RAF kinase domain bound to the inhibitor 2a | | Descriptor: | 1-(propan-2-yl)-3-({3-[3-(trifluoromethyl)phenyl]isoquinolin-8-yl}ethynyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Kurinov, I, Assadieskandar, A, Yu, C, Liu, X, Chen, Y.-C, Prakash, G.K.S, Zhang, C, SIcheri, F. | | Deposit date: | 2018-01-30 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Effects of rigidity on the selectivity of protein kinase inhibitors.
Eur J Med Chem, 146, 2018
|
|
8WTQ
 
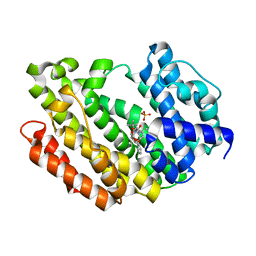 | | HUMAN SQUALENE SYNTHASE IN COMPLEX WITH {1-[2-Chloro-5-(2,2-dimethyl-propyl)-13-(2-methoxy-phenyl)-6-oxo-6,7,10,11-tetrahydro-5H,9H,13H-12-oxa-5,8-diaza-benzocycloundecene-8-carbonyl]-piperidin-4-yl}-acetic acid | | Descriptor: | 2-[1-[[(10~{S})-13-chloranyl-2-(2,2-dimethylpropyl)-10-(2-methoxyphenyl)-3-oxidanylidene-9-oxa-2,5-diazabicyclo[9.4.0]pentadeca-1(15),11,13-trien-5-yl]carbonyl]piperidin-4-yl]ethanoic acid, PHOSPHATE ION, Squalene synthase | | Authors: | Suzuki, M, Haginoya, N, Suzuki, M, Ishigai, Y, Terayama, K, Kanda, A, Sugita, K. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel 11-Membered Templates as Squalene Synthase Inhibitors.
J.Med.Chem., 67, 2024
|
|
8WTR
 
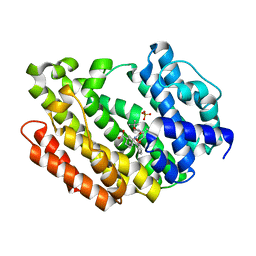 | | HUMAN SQUALENE SYNTHASE IN COMPLEX WITH (1S,3R)-3-[2-Chloro-5-(2,2-dimethyl-propyl)-13-(2-methoxy-phenyl)-6,8-dioxo-5,6,7,8,10,11-hexahydro-13H-12-oxa-5,9-diaza-benzocycloundecen-9-yl]-cyclohexanecarboxylic acid | | Descriptor: | (1~{S},3~{R})-3-[(10~{S})-13-chloranyl-2-(2,2-dimethylpropyl)-10-(2-methoxyphenyl)-3,5-bis(oxidanylidene)-9-oxa-2,6-diazabicyclo[9.4.0]pentadeca-1(15),11,13-trien-6-yl]cyclohexane-1-carboxylic acid, PHOSPHATE ION, Squalene synthase | | Authors: | Suzuki, M, Haginoya, N, Suzuki, M, Ishigai, Y, Terayama, K, Kanda, A, Sugita, K. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel 11-Membered Templates as Squalene Synthase Inhibitors.
J.Med.Chem., 67, 2024
|
|
8CC4
 
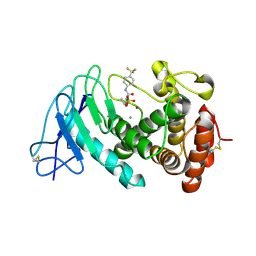 | | LasB bound to phosphonic acid based inhibitor | | Descriptor: | CALCIUM ION, Elastase, ZINC ION, ... | | Authors: | Mueller, R, Sikandar, A. | | Deposit date: | 2023-01-26 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibitors of the Elastase LasB for the Treatment of Pseudomonas aeruginosa Lung Infections.
Acs Cent.Sci., 9, 2023
|
|
5W3K
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase in complex NADPH, Mg2+ and CPD | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Patel, K.M, Teran, D, Zheng, S, Kandale, A, Schembri, M, McGeary, R.P, Schenk, G, Guddat, L.W. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Crystal Structures of Staphylococcus aureus Ketol-Acid Reductoisomerase in Complex with Two Transition State Analogues that Have Biocidal Activity.
Chemistry, 23, 2017
|
|
7OC7
 
 | | LasB, alpha-alkyl-N-aryl mercaptoacetamide | | Descriptor: | (2R)-N,3-diphenyl-2-sulfanyl-propanamide, CALCIUM ION, Neutral metalloproteinase, ... | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2021-04-26 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate-Inspired Fragment Merging and Growing Affords Efficacious LasB Inhibitors.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5O28
 
 | | E. coli FolD apo | | Descriptor: | Bifunctional protein FolD | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2017-05-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The natural product carolacton inhibits folate-dependent C1 metabolism by targeting FolD/MTHFD.
Nat Commun, 8, 2017
|
|
7NLM
 
 | | LasB, N-aryl-2-butylmercaptoacetamide | | Descriptor: | (S)-2-mercapto-N-(4-methoxyphenyl)-4-methylpentanamide, CALCIUM ION, Pro-elastase, ... | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2021-02-22 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | LasB, N-aryl-2-butylmercaptoacetamide inhibitor complex
to be published
|
|
